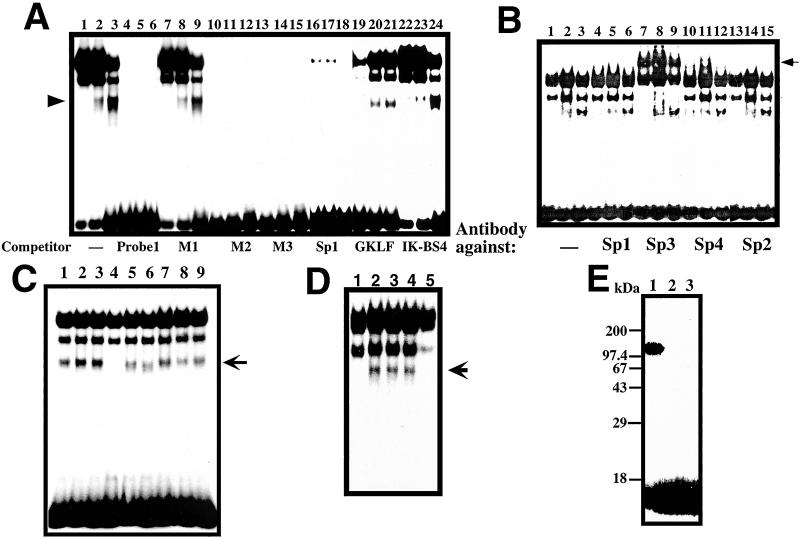
Figure 3.
GMSA analysis of Reg gene promoter. (A) Binding of RINm5F cell nuclear extracts to the cis-element by GMSA. Nuclear extracts from untreated cells were applied onto lanes 1, 4, 7, 10, 13, 16, 19, and 22; those from IL-6/dexamethasone (Dx)-treated cells were applied onto lanes 2, 5, 8, 11, 14, 17, 20, and 23; and those from IL-6/Dx/nicotinamide (NA)-treated cells were applied onto lanes 3, 6, 9, 12, 15, 18, 21, and 24. (B) Supershift assay of the cis-element binding factors by antibodies against Sp1 (lanes 4–6), Sp2 (lanes 13–15), Sp3 (lanes 7–9), and Sp4 (lanes 10–12). Nuclear extracts from untreated cells were applied onto lanes 1, 4, 7, 10, and 13; those from IL-6/Dx-treated cells were applied onto lanes 2, 5, 8, 11, and 14; and those from IL-6/Dx/NA-treated cells were applied onto lanes 3, 6, 9, 12, and 15. (C) Effects of poly(ADP-ribosyl)ation of nuclear proteins on the binding ability to the cis-element by GMSA. 0.2 μl of 100 mM β-NAD+, 100 mM α-NAD+, 100 mM ADP-ribose, 1 mM cyclic ADP-ribose, 100 mM NA, and/or 10 mM 3-aminobenzamide (3AB) were added in a 20-μl reaction before the addition of the 32P-labeled probe 1. Lane 1, none; lane 2, NA; lane 3, 3AB; lane 4, β-NAD+; lane 5, β-NAD++NA; lane 6, β-NAD++3AB; lane 7, α-NAD+; lane 8, ADP-ribose; and lane 9, cyclic ADP-ribose. (D) The involvement of PARP in the active transcriptional complex was evidenced by the immunodepletion of PARP. An active transcriptional complex of the nuclear extract from RINm5F cells stimulated by IL-6/Dx (lane 2) was blocked by the addition of 1 mM β-NAD+ as described above (lane 1).The complex formation was inhibited by the treatment of nuclear extract with an anti-PARP antibody (lane 5) but not with an anti-CD38 antibody (lane 4) nor with preimmune serum (lane 3). An arrow indicates the position of the active transcriptional complex. (E) Evidence of autopoly(ADP-ribosyl)ation of PARP in the GMSA reaction. Poly(ADP-ribosyl)ation was performed in the same condition as GMSA in the presence of 62 nM [adenylate-32P]NAD+ (2 × 106 cpm, NEN). After the incubation, proteins were precipitated by the addition of 9-volume acetone, separated by SDS/PAGE (12.5%), and autoradiographed. NA (1 mM, lane 2) or 3AB (0.1 mM, lane 3) was added in the GMSA incubation.