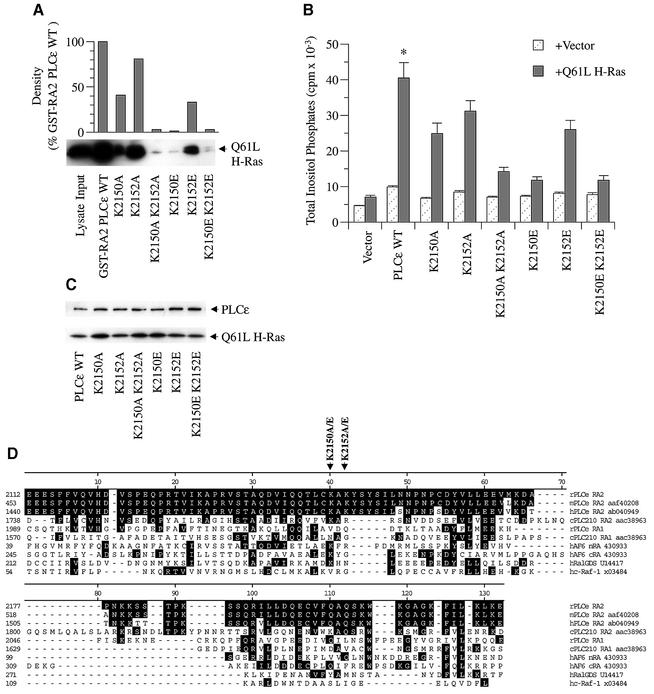
Fig. 7. Effect of specific RA2 mutations on Ras binding and activation of PLCε. (A) Binding of activated H-Ras to wild-type and mutant PLCε RA2 domains. Mutations in the RA2 domain of GST–RA2 PLCε as shown in (D) were introduced by site-directed mutagenesis. Binding was determined by GST pull-down assays using lysate from COS-7 cells overexpressing constitutively active Q61L H-Ras (input) as detailed in Figure 3 and Materials and methods. An image of an autoradiograph of a western blot stained with anti-Ras (arrow) is shown. Upper panel: densitometry of multiple exposures and dilutions was performed to quantitate autoradiographs. (B) Effect of RA2 mutations on PLCε activation by activated Ras. COS-7 cells were transiently transfected with 0.5 µg of vector (pCMV-LacZ), wild-type PLCε (pCMV-PLCε-FLAG), or PLCε with the indicated RA2 mutation and vector (pRSV-LacZ) or Q61L H-Ras (pRSV-Leu61). Values are the mean ± SEM of four experiments performed in duplicate. *P at least <0.016 compared with all mutants + Q61L H-Ras. (C) Expression of the indicated construct in the presence of Q61L H-Ras. A western blot was stained concurrently with FLAG (PLCε) and Ras (Q61L H-Ras) antibodies as indicated by arrows. The blot is representative of three similar experiments. (D) Alignment of the Ras-binding domains of rat PLCε (rPLCε af323615), mouse PLCε (mPLCε aaf40208), human PLCε (hPLCε ab040949), C.elegans PLC210 (cPLC210 aac38963), human AF6 (hAF6 430933), human RalGDS (hRalGDS u14417) and human c-Raf-1 (hc-Raf-1 x03484). Clustal alignment was performed with MegAlign (DNASTAR, Madison, WI) and adjusted manually. Black shading represents amino acid identity with rat PLCε. The locations of amino acid mutations are indicated by arrows. K2150 and K2152 (lysines at position 2150 and 2152) were mutated to A (alanine) or E (glutamic acid).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.