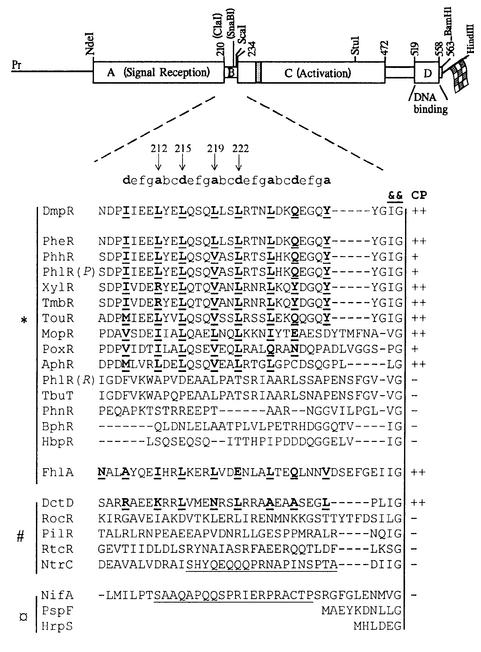
Fig. 1. Coils analysis of the linker region of DmpR and other σ54-dependent transcriptional regulators. Above, schematic representation of dmpR indicating the location of restriction sites used in generating DmpR derivatives. The domain structure of DmpR, discussed in the text, is superimposed on the restriction map. The shaded box indicates the location of the ATP-binding motif of the family (Morett and Segovia, 1993) and the flag represents a C-terminal fusion of an eight amino acid epitope tag. Below, alignment of the linker region of selected members of the σ54-dependent family of transcriptional activators. Alignment was made relative to two highly conserved residues, a hydrophobic residue followed by glycine (indicated by &&), which mark the start of the C-domain (Jaspers et al., 2000). The a- and d-positions within heptad (a-b-c-d-e-f-g) repeats of proteins predicted to form a coiled coil by the Coils program (Lupas et al., 1991) are shown in bold and underlined. Arrows indicate residues of DmpR targeted in specific mutants used in this study. ‘CP’ indicates the propensity prediction to form a coiled coil of >90% (++), >50% (+) or <50% (–). The linker regions of the different family members are grouped according to known or suspected activation mechanisms with * indicating small-ligand responsive regulators, # indicating phosphorylation-responsive regulators, and ¤ indicating those lacking an A-domain and/or controlled by protein–protein interactions. Underlined sequence indicates the Q-linker regions of NtrC and NifA referred to in the text. Sequences are: AphR from Comamonas testoseroni TA441 (X68033; Arai et al., 1998); BphR encoded on plasmid pNL1 from Sphingomonas aromaticivorans F199 (AF079317; Romine et al., 1999); DctD from Rhizobium meliloti (P13632; Watson, 1990); DmpR encoded on plasmid pVI150 from Pseudomonas sp. strain CF600 (A47078; Shingler et al., 1993); FhlA from E.coli K-12 (P19323; Schlensog and Bock, 1990); HbpR from Pseudomonas azelaica HBP1 (U73900; Jaspers et al., 2000); HrpS from Pseudomonas syringae (P26316; Xiao et al., 1994); MopR from Acinetobacter calcoaceticus NCIB8250 (Z69251; Schirmer et al., 1997); NifA from Klebsiella pneumoniae (P03027; Arnold et al., 1988); NtrC from K.pneumoniae (P03029; Drummond et al., 1983); PheR from Pseudomonas putida BH (D63814; M.Takeo, unpublished); PhhR from P.putida P35X (X79599; Ng et al., 1995); PhlR encoded on plasmid pPGH1 from P.putida H (Müller et al., 1996) indicated in the figure as PhlR(P); PhlR from Ralstonia eutropha JMP134 (AF065891; Ayoubi and Harker, 1998) indicated in the figure as PhlR(R); PhnR from Burkolderia sp. strain RP007 (AF061751; Laurie and Lloyd-Jones, 1999); PilR from Pseudomonas aeruginosa PA01 (Q00934; Ishimoto and Lory, 1992); PoxR from R.eutropha E2 (GAF026065; Hino et al., 1998); PspF from E.coli K-12 (P37344; Jovanovic et al., 1997); RocR from Bacillus subtilis (P38022; Calogero et al., 1994); RtcR from E.coli K-12 (P38035; Blattner et al., 1997); TbuT from Ralstonia pickettii PK01 (U72645; Byrne and Olsen, 1996); TmbR from P.putida TMB (U41301; Favaro et al., 1996); TouR from P.putida (AJ005663; Bertoni et al., 1998); and XylR encoded on plasmid pWW0 from P.putida mt-2 (M10143; Inouye et al., 1985).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.