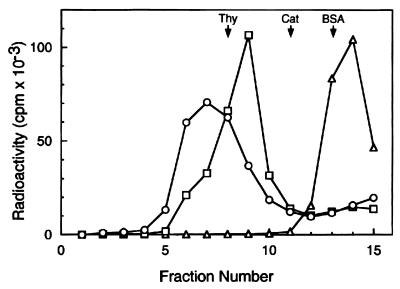
Figure 4.
Glycerol gradient sedimentation analysis of Mcm/DNA complexes. Mcm4/6/7 complex was incubated with 0.5 pmol of a 32P-labeled DNA substrate containing a 60-nt 3′ tail and a 30-nt 5′ tail with a 50-bp duplex region (the same substrate described in Fig. 3A) in a reaction mixture (150 μl) containing 25 mM Hepes-NaOH, pH 7.5, 50 mM sodium acetate, 10 mM magnesium acetate, 1 mM DTT, and 0.1 mg/ml BSA. After incubation at 25°C for 30 min, the reaction mixture was loaded onto a 5-ml 15–35% glycerol gradient in a buffer A containing 0.1 mg/ml BSA. After centrifugation at 48,000 rpm for 6 h in a Beckman SW 50.1 rotor at 4°C, fractions (330 μl) were collected from the bottom of the tube. The distribution of Mcm/DNA complexes or DNA were determined by liquid scintillation counting. The marker proteins used were thyroglobulin (Thy, 19S), catalase (Cat, 11.3S), and BSA (BSA, 4.3S). ○, The binding reaction was carried out with 7.5 μg of the Mcm4/6/7 complex (13 pmol Mcm4/6/7 dimer) to assemble the C2 complex; □, 0.3 μg of the Mcm4/6/7 complex was used (about 0.5 pmol Mcm4/6/7 dimer) for the formation of the C1 complex; ◊, DNA substrate alone.