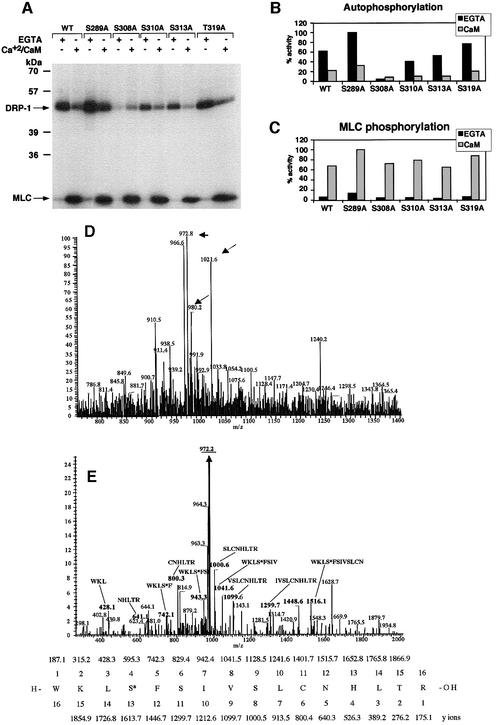
Fig. 3. Identification of Ser308 as the major autophosphorylation site of DRP-1 in vitro. (A) DRP-1 and its point mutation constructs were expressed and imunoprecipitated with anti-HA antibodies. The proteins were then assayed in vitro for kinase activity (autophosphorylation and MLC phosphorylation) in the presence or absence of Ca2+/CaM as described in Figure 1A. (B and C) Bars showing the relative amount of autophosphorylation and MLC phosphorylation by DRP-1 point mutations in the presence or absence of Ca2+/CaM, after normalizing to protein expression levels as described in Figure 1C and D. (D and E) Mapping autophosphorylation sites by liquid chromatography–mass spectrometry. (D) Full mass spectrum at retention time 53 min. The masses corresponding to peptide 305–320 in the unphosphorylated (m/z = 980.2), the phosphorylated (m/z = 1021.6) and the dephosphorylated forms (m/z = 972.8) are marked by arrows. (E) Collision-induced dissociation (CID) of the m/z = 1021.6 peptide and comparison with the simulated CID of phosphorylated 305–320 DRP-1 peptide. The major fragment m/z = 972.2 (underlined) correlates with the doubly charged mass of the full peptide after H3PO4 removal (indicating the existence of a single phosphate residue on this peptide). The relevant CID fragments are marked in bold and their amino acid sequence and mass are shown. It indicates that the phosphate residue resides exclusively on Ser308 and not on Ser310, Ser313 or Thr319. The CID simulation of the phosphorylated 305–320 DRP-1 peptide is shown at the bottom. The phosphoserine is marked as S*.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.