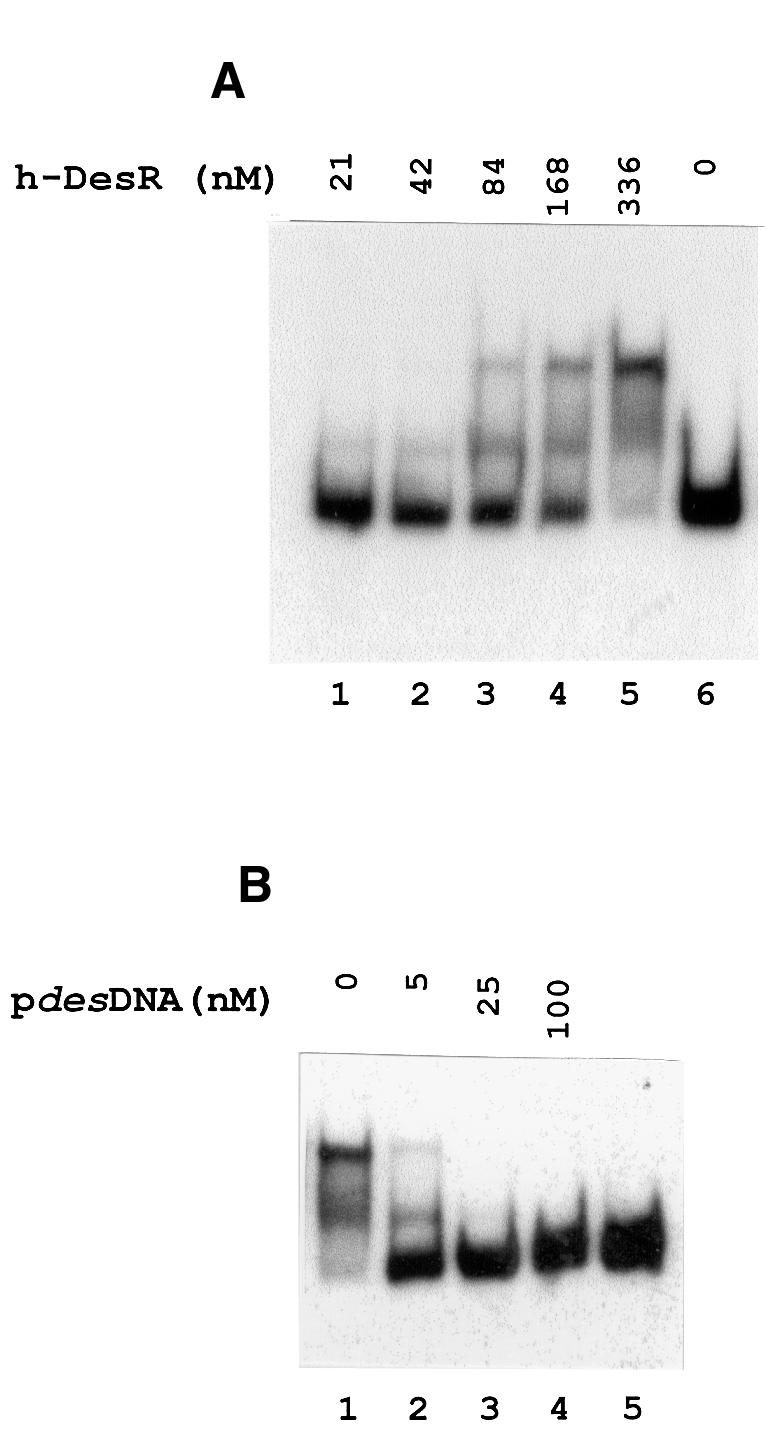
Fig. 3. Gel shift assay showing the binding of DesR to the des promoter region. (A) The 367 bp des promoter fragment (pdesDNA) was prepared by [α-32P]dATP PCR labelling as described in Materials and methods. The [32P]pdesDNA concentration in the binding mixtures was 1.7 nM in all cases. The concentration of h-DesR used in each binding reaction is indicated above the respective line. (B) Specific competition in binding reactions using 1.7 nM [32P]pdesDNA and 336 nM h-DesR. Lane 1 shows the retarded species in the absence of unlabelled homologous DNA. Lanes 2, 3 and 4 show the dissociation of the labelled complex in the presence of 3, 15 and 60-fold molar excess of unlabelled pdesDNA, respectively, added to the binding mixtures before the addition of h-DesR. Lane 5 shows [32P]pdesDNA without the addition of h-DesR.
