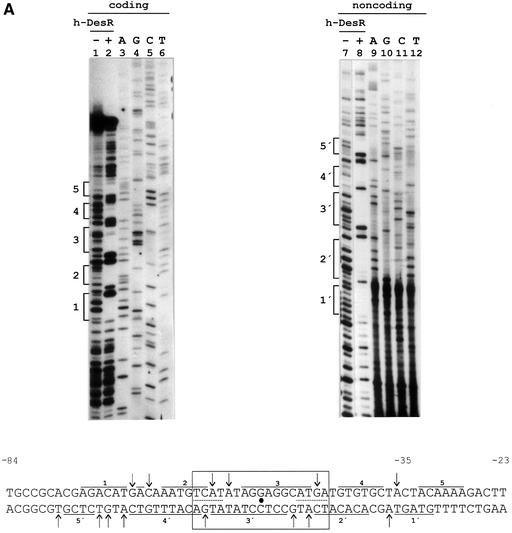
Fig. 4. DNase I footprinting assay of the des promoter region and in vivo characterization of the des promoter protected region. (A) DNase I footprinting of h-DesR protein on both strands of a 178 bp DNA fragment containing the des promoter (see Materials and methods). Sequencing reactions were performed on the same DNA fragment labelled at the coding (lanes 3 to 6) and non-coding (lanes 9 to 12) strands. Lanes 1, 2, 7 and 8 show the DNase I digestion products of pdesDNA in the presence (+) or absence (–) of h-DesR. Brackets mark the protected regions in each strand. The putative 17 bp symmetric region of the protected region is boxed, with the dyad axis of symmetry indicated by a dot. The inverted repeats are underlined with dots. DNase I footprints on both strands are shown. Arrows indicate hypersensitive bonds. (B) Promoter mutations. The sequence changes in the promoter variants are depicted along the protected region of the des promoter. The deleted region is indicated by dots, and the nucleotide changes to introduce mutations in the left inverted repeat are shown in bold characters. The inverted repeat sequences are underlined. The strains were grown at 37°C to an OD of 0.30 and then subjected to a downshift to 25°C. After 3 h of growth at 25°C, the cells were harvested and β-gal activities were determined. The average value of β-gal activity of strain AKP3 (bearing the wild-type promoter) was taken as 100% of promoter activity. The results shown are the average of three independent experiments.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.