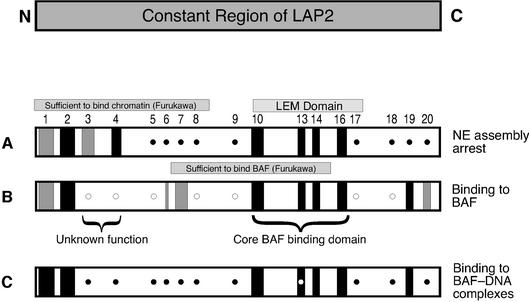
Fig. 5. Summary of mutagenesis results for Xenopus LAP2-c. Numbers indicate the position of each cluster of mutations in the LAP2 amino acid sequence (see Figure 1). Each row summarizes the activity of the mutant proteins in a given assay, as described in the text. (A) ability to inhibit nuclear assembly in Xenopus egg extracts; (B) ability to bind to BAF; (C) ability to supershift BAF⋅DNA complexes. Closed circles indicate that the mutant protein had the same activity as wild-type LAP2-c. Open circles indicate that the mutant protein had more activity (e.g. higher affinity for BAF) than wild-type LAP2-c. Black bars indicate loss of activity. Gray bars indicate partial loss of activity. An open circle inside a black bar for mutant m13 indicates that this mutant bound to BAF⋅DNA complexes in six out of 10 experiments. The LEM domain is indicated, as well as regions in Xenopus LAP2 that correspond to those sufficient to bind BAF in two-hybrid assays (rat residues 67–137; Furukawa, 1999), or sufficient to bind chromatin in vitro (rat residues 1–88; Furukawa et al., 1997).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.