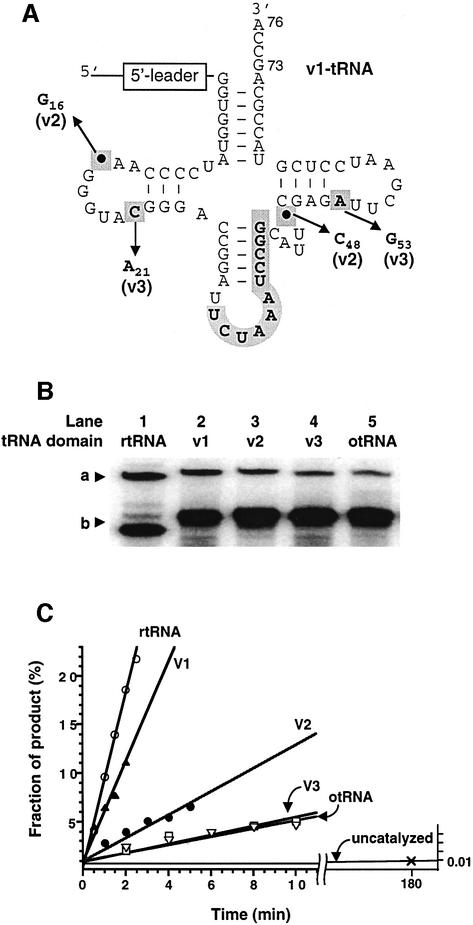
Fig. 5. Structure and self-aminoacylation activity of pre-24 and its variants. (A) Structure of otRNA variants introduced in the otRNA domain of pre-24. The mutations and deletions observed in rtRNA are highlighted by bold letters in gray boxes (see also Figure 1B). In order to distinguish the base numbering of tRNA from that of the 5′-leader domain, the subscript number is used for the assignment of tRNA bases according to the tRNA numbering rule (see Figure 3B). The structure of each variant is as follows: v1-tRNA (its structure is shown), in which the original anticodon region of otRNA was restored but the point mutations and deletions observed in rtRNA were maintained; v2-tRNA, in which the two point deletions were restored into v1-tRNA but the mutations were maintained; v3-tRNA, in which the two point mutations were restored into v1-tRNA but the deletions were maintained. (B) Comparison of self-aminoacylation activity of pre-24 and its variants with different tRNA domains. a, Biotin-Phe-pre-24 or variants complexed with SAv; b, pre-24 or variants. Reactions were carried out in the presence of 1 µM pre-24 (lane 1), variants (lanes 2–4) or pre-24otRNA (lane 5), incubating with 1 mM Biotin-Phe-CME at 25°C for 2 h. (C) Initial rates of self-aminoacylation of pre-24 and its variants. Reactions were carried out in the presence of 0.5 µM pre-24 (rtRNA) or its variants (v1–v3-tRNA and otRNA) incubated with 5 mM Biotin-Phe-CME at 25°C. The observed rate constants (kobs) are 0.089 min–1 (rtRNA), 0.051 min–1 (v1-tRNA), 0.012 min–1 (v2-tRNA), 0.0047 min–1 (v3-tRNA) and 0.0043 min–1 (otRNA).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.