Abstract
Replicative helicases are motor proteins that unwind DNA at replication forks. Escherichia coli DnaB is the best characterized member of this family of enzymes. We present the 26 Å resolution three-dimensional structure of the DnaB hexamer in complex with its loading partner, DnaC, obtained from cryo-electron microscopy. Analysis of the volume brings insight into the elaborate way the two proteins interact, and provides a structural basis for control of the symmetry state and inactivation of the helicase by DnaC. The complex is arranged on the basis of interactions among DnaC and DnaB dimers. DnaC monomers are observed for the first time to arrange as three dumb-bell-shaped dimers that interlock into one of the faces of the helicase. This could be responsible for the freezing of DnaB in a C3 architecture by its loading partner. The central channel of the helicase is almost occluded near the end opposite to DnaC, such that even single-stranded DNA could not pass through. We propose that the DnaB N-terminal domain is located at this face.
Keywords: cryo-electron microscopy/DNA replication/helicases/molecular motors/three-dimensional reconstruction
Introduction
Strand separation in DNA duplexes is a key step in many cellular events. Helicases are the motor proteins responsible for DNA unwinding, a process fuelled by the hydrolysis of a nucleoside triphosphate (Lohman and Bjornson, 1996; Patel and Picha, 2000). The unwinding step occurs with a specific polarity with respect to the strand on which the particular helicase translocates.
DnaB is the major replicative helicase in Escherichia coli. It is the best characterized member of the family of hexameric replicative helicases that associate with primases (Patel and Picha, 2000). To date, high-resolution structural information about the hexameric helicases is restricted to the gene 4 helicase of bacteriophage T7 (Sawaya et al., 1999; Singleton et al., 2000) and the small N-terminal domain of DnaB (Fass et al., 1999; Weigelt et al., 1999).
Replicative helicases interact with other proteins of the replication machinery in larger protein assemblies, such as the replisome, which coordinates both simultaneous DNA synthesis on the leading and lagging strands at a replication fork and fork movement (Baker and Bell, 1998). The role of helicases in replication thus extends far beyond the unwinding of duplex DNA. A complete understanding of these molecular motors will necessarily include the structural and functional characterization of their complexes with other proteins. DnaB, for instance, interacts with the DnaG primase (Lu et al., 1996; Chang and Marians, 2000), as well as the τ subunit of the DNA polymerase III holoenzyme (Kim et al., 1996), and the replication terminator protein RTP (Manna et al., 1996).
Initiation of chromosomal replication also requires DnaB to interact with its loading partner DnaC, which properly delivers the helicase to a DnaA-containing nucleoprotein complex at the origin of replication oriC (Baker et al., 1986). Native DnaB is a homohexamer of 52-kDa monomers that, in the presence of ATP, forms a complex with six 28-kDa monomers of DnaC (Wickner and Hurwitz, 1975; Kobori and Kornberg, 1982b). The DnaB·DnaC complex is not itself active as a helicase (Wahle et al., 1989b). Transfer of DnaB to single-stranded (ssDNA), which presumably requires ATP hydrolysis, is accompanied by release of DnaC (Funnell et al., 1987; Wahle et al., 1989a). After having been loaded, DnaB begins DNA unwinding with 5′ to 3′ polarity.
Earlier cryo-electron microscopy (cryo-EM) studies of DnaB·DnaC that had been diluted into buffer containing ADP (San Martín et al., 1998) provided the only available three-dimensional (3D) reconstruction of a helicase in a complex with an auxiliary protein. Nevertheless, the limited resolution achieved (42 Å) imposed severe constraints on the interpretation of the reconstructed volume. In this work, we have obtained the DnaB·DnaC structure at considerably higher resolution (26 Å), using cryo-EM of samples in buffer containing ATP, to ensure the stability of the complex. The results allow for a much deeper insight into this macromolecular assembly and also reveal novel features that have important functional implications.
Results and discussion
Electron microscopy and image analysis of the DnaB·DnaC complex
Vitrified samples of purified DnaB·DnaC complex showed different views of the particle when imaged at 0° tilt (Figure 1A). Along with the previously described ring-like top view of the complex (San Martín et al., 1998) a wider range of views were found, including a minor proportion of side views. Micrographs obtained at different tilt angles (35° maximum) guaranteed a more even angular distribution of views (Figure 1B). Some 8000 particles were manually extracted and included in the reconstruction scheme; a gallery of these images is shown in Figure 1C.
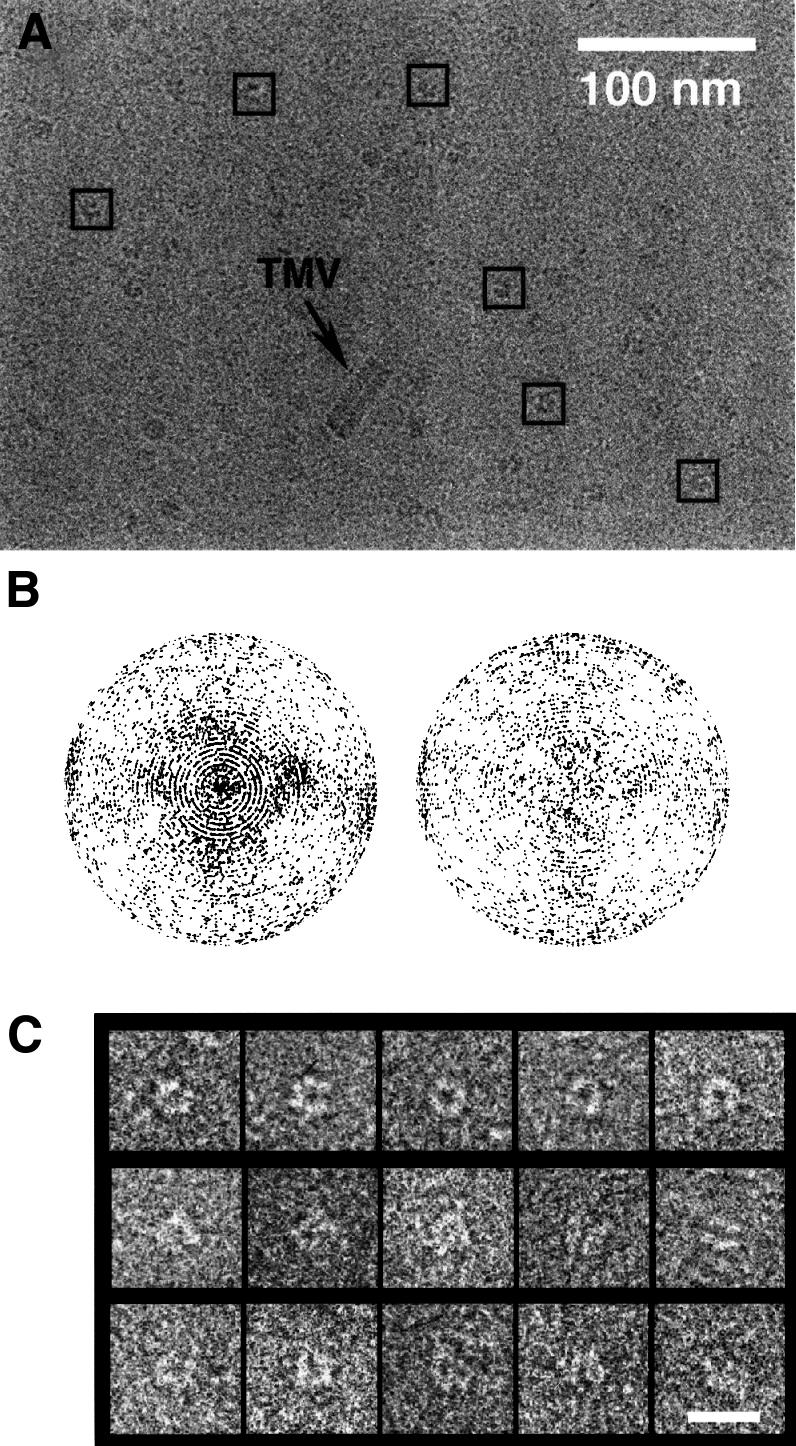
Fig. 1. Cryo-EM of the DnaB·DnaC complex. (A) Electron micrograph of a vitrified preparation of E.coli DnaB·DnaC imaged at 0° tilt. Some particles selected from this micrograph are indicated by squares. (B) Angular distribution of the projection images from untilted (left) and tilted (right) micrographs. Each projection image is represented by a point in a sphere, viewed in the direction of the polar axis. Pure top views are at the pole of the hemisphere, whereas pure side views are on the equator. The longitude of each point represents the azimuthal angle of the projection image. (C) Galleries of top views (first row), side views (second row) and intermediate views (third row). The bar represents 20 nm.
3D reconstruction of DnaB·DnaC
After angular assignment and refinement of the projection images, a 3D electron density map of the DnaB·DnaC complex was calculated (see Materials and methods). The 3D reconstruction presented in Figure 2 has a resolution of 26 Å. In accordance with the earlier results, the reconstructed volume has a general toroidal shape. However, the inner channel is far from being homogeneous in size; it narrows considerably at one of the apical faces of the volume.
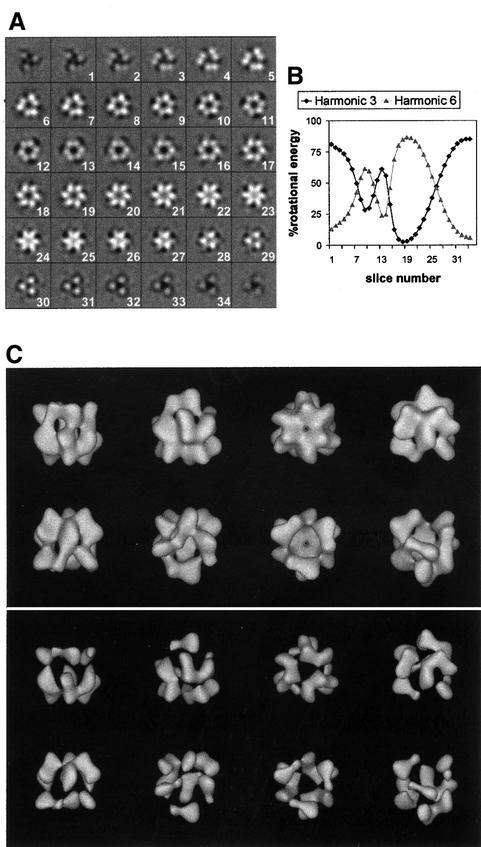
Fig. 2. DnaB·DnaC complex at 26 Å resolution. (A) Slices through the reconstructed volume. Significant slices extend from 1 to 34. (B) Variations in symmetry along the rotational axis of the volume. The plots represent the relative energy of the 3- and 6-fold components of the rotational power spectra of successive slices. (C) Surface renderings of the volume at a threshold accounting for 100% (top) and 40% (bottom) of the expected mass. The volume was symmetrized and filtered to its resolution.
The maximum external diameter of the particle is 13.8 nm and the reconstructed height is 12.4 nm. These dimensions are larger than those previously reported [12.5 and 9.1 nm, respectively (San Martín et al., 1998)]. Several factors explain these differences. Inclusion of a standard for magnification [tobacco mosaic virus (TMV)] allowed calculation of the real magnification and consequent correction of the voxel size, thus increasing the reliability of measurements of dimensions. Moreover, the random conical tilt reconstruction scheme used by San Martín et al. (1998) brings down the resolution in the Z-axis direction, while in the present study, the even angular distribution of projection images precludes potential artefacts that would arise from loss of information in some directions. Also, higher resolution renders smaller pieces of mass previously undetected (e.g. see the last slices in Figure 2A), which in turn increases the height to diameter ratio of the volume.
Assignment of DnaB and DnaC within the volume
The cryo-reconstruction of DnaB alone, calculated at 35 Å resolution by San Martín et al. (1998), was significantly shorter than the DnaB·DnaC volume in the direction along the axis of rotational symmetry. This indicated that DnaC most likely sits on one of the faces of the toroidal DnaB hexamer. In the new reconstruction, reliable localization of the helicase and its loading partner within the volume is feasible. On examination of the surface renderings shown in Figure 2C, it is apparent that the reconstructed volume contains two clearly distinct regions, one on top of the other. The first spans between slices 1 and 12 (approximately, in Figure 2A), and the second includes the last 22 slices of the 34 that are considered to bear significant information. Increasing the threshold to highlight the areas of maximum density leads to the two regions segmenting apart (Figure 2C). Both regions have a triangular shape defined by their 3-fold symmetry. However, one triangle is twisted relative to the other, which emphasizes the fact that they comprise different building blocks of the complex. Moreover, the two regions account for 65 and 35% of the reconstructed height, figures that match the relative masses of DnaB and DnaC, respectively, in the complex. These two regions, therefore, almost certainly correspond to the helicase and its loading partner.
This assignment is further supported by detailed examination of the symmetry of the complex. DnaC-free DnaB hexamers can adopt two different architectures of C3 and C6 symmetries, which co-exist under certain conditions (Yu et al., 1996; Donate et al., 2000). This DnaB polymorphism is pH dependent, and at pH 7.6 DnaB adopts only the C3 quaternary state (Donate et al., 2000). In the DnaB·DnaC complex, DnaB is always in the C3 architecture, regardless of the pH (Donate et al., 2000). In this work, therefore, DnaB is expected to adopt a C3 conformation both because of the pH used (7.6) and its liaison with DnaC. The 3D cryo-reconstruction of the C3 architecture of DnaB (San Martín et al., 1998) showed that the apical faces of the helicase differed in that one had a 3-fold and the other had a predominant 6-fold symmetry component. Examination of symmetry variations along the axis of rotational symmetry of the DnaB·DnaC volume (Figure 2B) shows that indeed there is a transition from 3- to 6-fold symmetry in the region considered to be DnaB, which supports the present assignment. Thus, the 3-fold symmetrical arrangement of DnaC molecules (discussed below) interacts with DnaB via the 6-fold face of the helicase. The interlocking of the two proteins would in part induce the observed fading of the 6-fold symmetry in the slices that correspond to the region of DnaB·DnaC contact (Figure 2A and B, slices 12–15).
Based on this assignment of regions of the reconstruction to DnaB and DnaC, there are novel interesting features with respect to the structures of the proteins and their interaction. These are summarized in Figures 3 and 4 and discussed next.
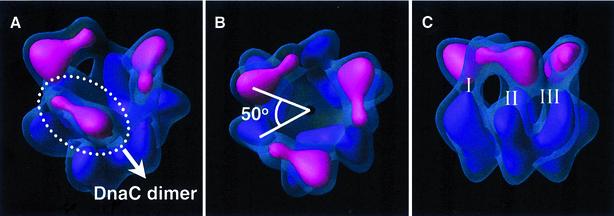
Fig. 3. DnaC in the complex. Surface renderings in semi-transparent blue account for 100% of the expected mass and those in opaque colours account for 40%. In the latter, the regions considered to be DnaC are depicted in pink, and the three DnaC dimers become segmented apart (A). There exists a relative twist among DnaC and DnaB dimers of ∼50° (measured as the relative rotation between the higher density areas of a DnaC dimer and a neighbouring DnaB dimer) (B). Three different contact areas (I, II and III) between each DnaC dimer and DnaB can be clearly distinguished (C).
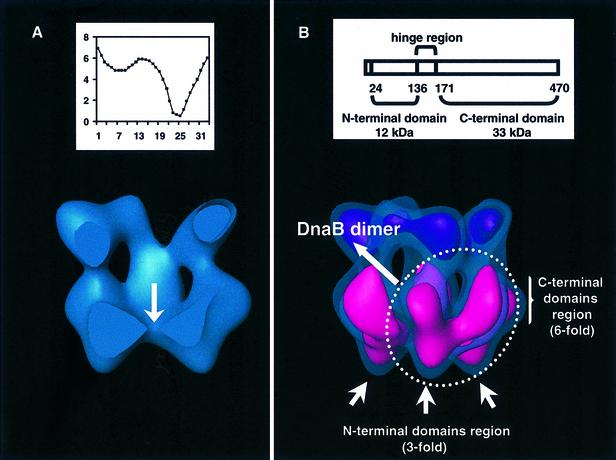
Fig. 4. DnaB in the complex. (A) Surface rendering representation of the DnaB·DnaC complex (100% of the mass) cut by a central plane through the length of the central channel. The arrow points towards the region where the channel narrows sharply. (Inset) Variations in the width of the central channel; the diameter of the channel (in nm) is plotted versus the slice number (Figure 2A). (B) Assignment of DnaB domains within the volume. The domain organization of the DnaB monomer is summarized in the scheme (inset), where numbers refer to residue positions in the primary structure. The reconstruction is presented at two different threshold values as in Figure 3. In this case, in the surface rendering that accounts for 40% of the mass, the region considered to be DnaB is highlighted in pink.
DnaC in the complex
Recruitment of the helicase to the fork assembly site is one of the primary goals in the initiation stage. In E.coli, DnaC is the protein responsible for the loading of DnaB at the chromosomal origin of replication. In some cases, such as in rolling-circle replication systems, helicase loading might not require a DnaC-type mechanism (Kornberg and Baker, 1992). Nevertheless, the assistance of a loading partner functionally equivalent to DnaC seems to be quite common to both prokaryotic and eukaryotic systems (Baker and Bell, 1998).
Loading partners may also maintain the helicases in an inactive state so that replication is not triggered until the replication machinery is properly recruited and the helicase–loading partner complex has disassembled. In E.coli, DnaC is released from the DnaB·DnaC complex via a hydrolysis-dependent reaction that specifically requires ATP (Funnell et al., 1987; Wahle et al., 1989a). Experiments carried out in the presence of ATP showed that DnaC can also have a stimulating effect on DnaB activities. This effect is indirect by promoting binding of the helicase to DNA. When ATP is replaced by ATPγS, all these activities are severely inhibited because the complex can no longer be disassembled (Wahle et al., 1989b).
Despite the known stoichiometry (6:1) of the DnaC– hexameric DnaB interaction (Kobori and Kornberg, 1982b), only three elongated masses appear in the DnaC region in the reconstructed complex presented here. Thus, DnaC appears to form three dimers when it interacts with DnaB. These dimers seem to be quite discrete; at the present level of resolution, clear regions of interaction between dimers are not detected (Figure 3).
This is the first indication of the existence of dimers of DnaC. All experimental data indicate that DnaC in solution exists only as a monomer (Kobori and Kornberg, 1982a), and that it binds either ATP or ADP (Galletto et al., 2000). The ATP- and ADP-bound forms of DnaC have different structures (Galletto et al., 2000), and NMR studies with 15N-labelled DnaC (in the absence of nucleotide) indicate that its structure is unusually flexible (G.Otting, S.E.Brown and N.E.Dixon, unpublished results). Taken together with the present results, these data suggest that DnaC undergoes structural changes on interaction with ATP and DnaB, and that these render it capable of forming DnaB-bound dimers that are now able to interact with DNA and other proteins to mediate loading of the helicase at origins of replication (Baker et al., 1986; Learn et al., 1997).
Furthermore, DnaC could thus control the quaternary state of the helicase by interacting with it in a dimeric fashion. If we assume that the C3/C6 polymorphism found in DnaB samples at physiological pH values is related in some way to conformational changes that occur during the enzymatic cycle of DnaB and/or its translocation on ssDNA during expression of its helicase activity, then the fact that it is frozen in the 3-fold architecture when bound to DnaC (Donate et al., 2000) provides a mechanism for its inactivation. This inactivation would prevent the use of its NTPase and helicase activities in inappropriate contexts (Learn et al., 1997).
Each of the putative DnaC dimers is dumb-bell shaped, with two regions of different size. Although DnaC dimers could be symmetrical (through the longest axis traversing them), their shape makes them interact asymmetrically with DnaB. In this regard, as a result of the relative twist (of ∼50°; Figure 3), DnaC interlocks into the DnaB hexamer, yielding extensive contacts between the two proteins, such that each DnaC dimer associates with two different dimeric units of DnaB. Three clearly defined areas of DnaB·DnaC interaction are distinguishable for every DnaC dimer: the smaller lobe of DnaC contacts one monomer of a DnaB subunit dimer (region I, Figure 3), while the larger lobe contacts both subunits of a neighbouring DnaB dimer (regions II and III, Figure 3).
DnaB in the complex
The helicase possesses overall 3-fold symmetry, implying an internal organization as a trimer of dimers. DnaB dimers are easy to locate within the structure at the present resolution. In particular, use of threshold values rendering <100% of the expected mass clearly segments them apart (Figures 2 and 4). DnaB dimers are strikingly asymmetrical, which means that the monomeric units adopt different orientations, or even different conformations, when assembled in the complex. There is a precedent for differences in the structure of two otherwise identical subunits in the case of the dimeric E.coli Rep helicase (Korolev et al., 1997).
The DnaB face on which DnaC sits has prominent 6-fold symmetry, wherein the six subunits of the helicase are clearly distinguishable. On the contrary, the opposite face of DnaB has 3-fold symmetry with three protruding lobes. Therefore, the quaternary structure of the helicase has remarkable polarity. Moreover, it also exhibits a prominent handedness in the 6-fold propeller-like region, undetected in the earlier 3D reconstruction. Polarity and handedness are common features among hexameric helicases, also being found with the T7 gene 4 helicase/primase (Egelman et al., 1995; Sawaya et al., 1999; Singleton et al., 2000) and the SV40 large T antigen (San Martín et al., 1997; Valle et al., 2000). Asymmetry might provide the structural basis for mechanisms that facilitate both proper loading of the helicase and polar DNA unwinding.
The inner channel through the particle narrows very conspicuously near the 3-fold end of DnaB (Figure 4A). In a surface rendering that accounts for 100% of the mass of the complex, the internal diameter of the molecule reaches a minimum size of ∼7 Å (slice 26, Figure 2A). Note that, due to limitations inherent to electron microscopy, the surface rendering for 100% of the mass corresponds to a median boundary within a shell of uncertainty. The channel is completely closed in surface renderings obtained with only slightly lower threshold values (not shown). Thus, at this level of resolution, we cannot be certain that the channel does not reach total closure. Nevertheless, it is clear that in a small region of the volume (between slices 23 and 28, approximately, in Figure 2A) the channel is too narrow to allow passage of even a single strand of DNA.
Nevertheless, DnaB by itself does bind ssDNA through this central channel (Jezewska et al., 1998b). This mode of binding is thought to be pivotal in its mechanism of action, and has also been proposed for other hexameric helicases (Egelman et al., 1995; Valle et al., 2000) and proven for T7 gene 4 helicase/primase (Hacker and Johnson, 1997) and DnaB from Thermus aquaticus (Kaplan, 2000). Thus, occlusion of the DnaB channel in the DnaB·DnaC complex is very likely a result of DnaC binding to the opposite face of the helicase molecule. The structure of a proteolytic fragment of the T7 gene 4 protein reveals the presence of three loop regions in the vicinity of the channel (Singleton et al., 2000). Analogous flexible regions in DnaB could participate in narrowing of the channel induced by the binding of DnaC. DnaB is inactive as a helicase while bound to DnaC (Wahle et al., 1989a); the constriction of the central channel in the DnaB·DnaC complex visualized here very likely contributes to helicase inactivation.
Location of domains of DnaB
The domains of DnaB may be tentatively located in the reconstructed volume. DnaB has two major compact regions: the N-terminal (12 kDa) and C-terminal (33 kDa) domains, linked by a flexible hinge region of ∼40 residues (Nakayama et al., 1984; Miles et al., 1997; Figure 4B). The C-terminal domain contains the sites for ssDNA binding and NTP hydrolysis, and it retains the capacity to hexamerize.
In negative stain images, the C3 form of DnaB appears to be built up of six similarly large masses arranged in a central ring, from which three smaller lobes protrude (San Martín et al., 1995; Yu et al., 1996; Donate et al., 2000). The C-terminal domains were proposed to correspond to the six larger central masses, whereas the three external lobes were assumed to be dimers of the N-terminal domains. The symmetry switch between C6 and C3 architectures in the intact helicase was proposed to occur via dimerization of N-terminal domains in the C3 state, which would separate in the C6 architecture, in a transition involving structural changes in the flexible interdomain hinge region. Structures of N-terminal domain fragments of DnaB at atomic resolution have recently been determined both by NMR (Weigelt et al., 1999) and X-ray crystallography (Fass et al., 1999). These studies showed that isolated N-terminal domains form symmetrical dimers of a defined structure and that a monomer–dimer equilibrium exists in solution, providing support for the central role of this domain in the symmetry transition.
These three external masses seen in negatively stained C3 DnaB images were not found in the cryo-EM reconstructions of either the DnaB·DnaC complex or DnaB alone (San Martín et al., 1998). In the present reconstruction of the DnaB·DnaC complex at 26 Å resolution, three lobes do appear at the 3-fold face of DnaB but protrude axially instead of radially. Therefore, the particular arrangement observed in the negatively stained images seems likely to be a consequence of deformations of the particle during the collapse induced by staining and dehydration. The plasticity of DnaB shown by the large-scale movements underlying the C3/C6 quaternary transition might make the helicase quite sensitive to such deformation.
Consequently, the N-terminal domains would be located at the 3-fold face of DnaB, whereas the C-terminal domains, which contain the primary hexamerization sites, would be located in the part of DnaB possessing the strongest 6-fold symmetry, where six lobes appear (Figure 4B). This type of assembly agrees with the suggestion that the N-terminal domains face away from the apex of the replication fork on the lagging strand [i.e. towards the 5′ arm (Jezewska et al., 1998a)].
There is growing evidence that the C-terminal domains also play an active role in the C6/C3 transition. The hexameric replicative helicase RepA (Scherzinger et al., 1997; Bárcena, 2000), despite lacking N-terminal domains, also exhibits C3/C6 polymorphism, which implies reorientations or conformational changes of the C-terminal domains (Bárcena, 2000). The observed overall shape of DnaB in the reconstructed complex also lends support to a general reorientation of the whole of the subunit when it adopts C3 architecture. DnaB dimers are strongly asymmetrical, precluding the existence of a 2-fold axis. Therefore, the transition from C6 architecture, where all subunits must be parallel, has to involve both rotations of the N-terminal domains around the hinge region so that they can dimerize, and movements of the C-terminal domains so that the dimers arrange in the observed asymmetrical, V-shaped configuration. Until the structures of both the C3 and the C6 architectures of the free helicase are obtained at sufficiently high resolution, the nature of the transition will remain unclear.
No known activity has been detected for the N-terminal domain alone. Nevertheless, it is absolutely required for DnaB to be fully active: when it is removed, the remaining fragment is defective in DNA unwinding, DNA priming, and in protecting DnaC from inactivation by N-ethylmaleimide (Nakayama et al., 1984; Biswas et al., 1994). The N-terminal domain is therefore considered to be essential for interactions between the helicase and both DnaC and the DnaG primase. A site of DnaG interaction has been mapped to part of the N-terminal domain (Chang and Marians, 2000), as would be expected if DnaB were positioned in the replication fork as described by Jezewska et al. (1998a). However, on the basis of the present assignment of DnaB domains within the complex, DnaC would interact with the C- rather than the N-terminal domain of the helicase. This means that the role of the N-terminal domain in the formation of the DnaB·DnaC complex would be indirect, by assisting the helicase to adopt a conformation that enables it to bind to DnaC via the C-terminal domains.
It should be noted that neither the direct interaction of a discrete N- or C-terminal domain with DnaC, nor the dimerization of the N-terminal domains in the intact helicase has yet been demonstrated experimentally. The work reported here paves the way for future studies which will establish this and many other aspects of helicase action that remain to be elucidated.
Materials and methods
Purification of the DnaB·DnaC complex
DnaB and DnaC proteins were simultaneously overproduced as described (San Martín et al., 1995), except that the E.coli strain used was AN1459/pPS560/pLysS, and the LB medium was further supplemented with chloramphenicol (50 mg/l). The two proteins were partially purified through an anion-exchange chromatography step in buffer containing ADP, as described (San Martín et al., 1998). The DnaB·DnaC complex was then prepared by mixing fractions containing DnaB and DnaC, followed by dialysis of the mixture against 50 mM Tris–HCl pH 7.6, 25 mM NaCl, 5 mM MgCl2, 2 mM dithiothreitol, 0.1 mM ATP, 20% (v/v) glycerol (buffer A) and rechromatography on a column of DEAE–Fractogel. The DnaB·DnaC complex, prepared in this way, has been shown to contain six monomers of DnaC for each hexamer of DnaB (N.Dammerova and N.E.Dixon, unpublished results). Purified protein samples were stored at –70°C.
Cryo-EM
DnaB·DnaC samples were diluted to 30 µg/ml in glycerol-free buffer A. A polydisperse sample of TMV fragments, generously provided by Dr Nicolas Boisset, was added as a standard for magnification. Samples were adsorbed for 30 s onto holey grids freshly coated with a thin carbon layer. Immediately after blotting, they were vitrified by plunging them into liquid ethane. Preparations were visualized in a Philips CM120 electron microscope equipped with a Gatan cryostage and a TVIPS CCD camera at 100 and 120 kV acceleration voltages. Images were taken at tilt angles ranging from 0 to 35°, nominal magnification of 60 000× and defocus between 1.0 and 1.5 µm. Electron micrographs were recorded under low dose conditions on Kodak SO-163 film.
Two-dimensional (2D) analysis
Micrographs were digitized in a Zeiss SCAI scanner with raster size 7 µm. Images were interpolated down by binning to a final pixel size of 3.6 Å. Particle selection and 2D analysis of the images were performed using Xmipp (Marabini et al., 1996) and/or SPIDER (Frank et al., 1996) software packages. A total of 8046 single particle images were extracted from the micrographs and centred. After screening and selection, the number of particles was reduced to 7888.
3D reconstruction
A 3D Radon transform (RT)-based system run in an iterative way (Radermacher, 1994) was used to refine the centring and to define the respective orientations of the projection images. A scaled DnaB·DnaC volume (San Martín et al., 1998) was used as the first reference volume. In this initial step, an angular increment of 3° was used to calculate the RT to be compared: the 3D RT of the reference volume and the 2D RT of the projection images. The search for the correlation maxima that define the orientations of the projection images was first carried out in increments of 12° for the three Eulerian angles over the full angular range. Three iterations were carried out. The reference volume was updated at every step with the projection images in their assigned orientations, and reconstructions were calculated by inverse 3D RT (Radermacher, 1997; Lanzavecchia et al., 1999). For angular refinement, the total population of DnaB·DnaC images was divided into three equally populated subsets. Iteratively, one of the groups was used to generate a reference volume to refine the position and angles of the rest of the images. The RT was calculated in angular increments of 2°. The search was restricted first to ±18° around the previous values of the Eulerian angles and carried out in increments of 6°. Subsequently, it was restricted to ±8° with increments of 2°.
Resolution was assessed periodically by applying the Fourier Shell Correlation criterion (Saxton and Baumeister, 1982). The angular refinement process was stopped after 35 iterations, when no further increase in resolution was observed. The final 3D reconstruction was calculated using an improved ART (algebraic reconstruction techniques) algorithm (Marabini et al., 1998). Fourier Shell Correlation using a threshold value of five times noise determined the final resolution (26 Å). The volume was then filtered to the estimated final resolution and the 3-fold symmetry was applied. The isosurface renderings were displayed using WEB (Frank et al., 1996) and AMIRA (http://amira.zib.de). The DnaB·DnaC complex mass represented at a given threshold value was calculated assuming a mean protein density of 1.33 g/cm3 (Squire and Himmel, 1979).
Acknowledgments
Acknowledgements
The help of O.Llorca, C.San Martín, P.Moutel and C.O.Sánchez-Sorzano is acknowledged. This work was partially supported by the European Union (grant PL970106), by the Spanish Agency CICYT (grant BIO 98-0761 to J.M.C.), by a joint German/Spanish grant (HA 1997-0049, to J.M.C. and M.R.) and by the NSF (grant DBI9515518 to M.R.). M.B. was supported by a Spanish FPI fellowship, L.E.D. by a contract from CSIC, and S.E.B. by an Australian Postdoctoral Fellowship.
References
- Baker T.A. and Bell,S.P. (1998) Polymerases and the replisome: machines within machines. Cell, 92, 295–305. [DOI] [PubMed] [Google Scholar]
- Baker T.A., Sekimizu,K., Funnell,B.E. and Kornberg,A. (1986) Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell, 45, 53–64. [DOI] [PubMed] [Google Scholar]
- Bárcena M. (2000) Análisis estructural del polimorfismo cuaternario en las helicasas replicativas y del complejo DnaB⋅DnaC de Escherichia coli. PhD thesis, Universidad Autónoma de Madrid, Madrid, Spain.
- Biswas S.B., Chen,P.-H. and Biswas,E.E. (1994) Structure and function of Escherichia coli DnaB protein: role of the N-terminal domain in helicase activity. Biochemistry, 33, 11307–11314. [DOI] [PubMed] [Google Scholar]
- Chang P. and Marians,K.J. (2000) Identification of a region of Escherichia coli DnaB required for functional interaction with DnaG at the replication fork. J. Biol. Chem., 275, 26187–26195. [DOI] [PubMed] [Google Scholar]
- Donate L.E., Llorca,O., Bárcena,M., Brown,S.E., Dixon,N.E. and Carazo,J.M. (2000) pH-controlled quaternary states of hexameric DnaB helicase. J. Mol. Biol., 303, 383–393. [DOI] [PubMed] [Google Scholar]
- Egelman E.H., Yu,X., Wild,R., Hingorani,M.M. and Patel,S.S. (1995) Bacteriophage T7 helicase/primase proteins form rings around single-stranded DNA that suggest a general structure for hexameric helicases. Proc. Natl Acad. Sci. USA, 92, 3869–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass D., Bogden,C.E. and Berger,J.M. (1999) Crystal structure of the N-terminal domain of the DnaB hexameric helicase. Structure, 7, 691–698. [DOI] [PubMed] [Google Scholar]
- Frank J., Radermacher,M., Penczek,P., Zhu,J., Li,Y.H., Ladjadj,M. and Leith,A. (1996). SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol., 116, 190–199. [DOI] [PubMed] [Google Scholar]
- Funnell B.E., Baker,T.A. and Kornberg,A. (1987) In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J. Biol. Chem., 262, 10327–10334. [PubMed] [Google Scholar]
- Galletto R., Rajendran,S. and Bujalowski,W. (2000) Interactions of nucleotide cofactors with the Escherichia coli replication factor DnaC protein. Biochemistry, 39, 12959–12969. [DOI] [PubMed] [Google Scholar]
- Hacker K.J. and Johnson,K.A. (1997) A hexameric helicase encircles one DNA strand and excludes the other during DNA unwinding. Biochemistry, 36, 14080–14087. [DOI] [PubMed] [Google Scholar]
- Jezewska M.J., Rajendran,S., Bujalowska,D. and Bujalowski,W. (1998a) Does single-stranded DNA pass through the inner channel of the protein hexamer in the complex with the Escherichia coli DnaB helicase? Fluorescence energy transfer studies. J. Biol. Chem., 273, 10515–10529. [DOI] [PubMed] [Google Scholar]
- Jezewska M.J., Rajendran,S. and Bujalowski,W. (1998b) Complex of Escherichia coli primary replicative helicase DnaB protein with a replication fork: recognition and structure. Biochemistry, 37, 3116–3136. [DOI] [PubMed] [Google Scholar]
- Kaplan D.L. (2000) The 3′-tail of a forked-duplex sterically determines whether one or two DNA strands pass through the central channel of a replication-fork helicase. J. Mol. Biol., 301, 285–289. [DOI] [PubMed] [Google Scholar]
- Kim S.S., Dallmann,H.C., McHenry,C.S. and Marians,K.J. (1996) Coupling of a replicative polymerase and helicase: a τ–DnaB interaction mediates rapid replication fork movement. Cell, 84, 643–650. [DOI] [PubMed] [Google Scholar]
- Kobori J.A. and Kornberg,A. (1982a) The Escherichia coli dnaC gene product. II. Purification, physical properties, and role in replication. J. Biol. Chem., 257, 13763–13769. [PubMed] [Google Scholar]
- Kobori J.A. and Kornberg,A. (1982b) The Escherichia coli dnaC gene product. III. Properties of the dnaB–dnaC protein complex. J. Biol. Chem., 257, 13770–13775. [PubMed] [Google Scholar]
- Kornberg A. and Baker,T.A (1992) DNA Replication. W.H. Freeman and Co., New York, NY.
- Korolev S., Hsieh,J., Gauss,G.H., Lohman,T.M. and Waksman,G. (1997) Major domain swiveling revealed by the crystal structures of complexes of E.coli Rep helicase bound to single-stranded DNA and ADP. Cell, 90, 635–647. [DOI] [PubMed] [Google Scholar]
- Lanzavecchia S., Bellon,P.L. and Radermacher,M. (1999) Fast and accurate three-dimensional reconstruction from projections with random orientations via Radon transforms. J. Struct. Biol., 128, 152–164. [DOI] [PubMed] [Google Scholar]
- Learn B.A., Um,S.-J., Huang,L. and McMacken,R. (1997) Cryptic single-stranded DNA binding activities of the phage λ P and Escherichia coli DnaC replication initiator proteins facilitate the transfer of E.coli DnaB helicase onto DNA. Proc. Natl Acad. Sci. USA, 94, 1154–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman T.M. and Bjornson,K.P. (1996) Mechanisms of helicase-catalyzed DNA unwinding. Annu. Rev. Biochem., 65, 169–214. [DOI] [PubMed] [Google Scholar]
- Lu Y.-B., Ratnakar,P.V.A.L., Mohanty,B.K. and Bastia D. (1996) Direct physical interaction between DnaG primase and DnaB helicase of Escherichia coli is necessary for optimal synthesis of primer RNA. Proc. Natl Acad. Sci. USA, 93, 12902–12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manna A.C., Pai,K.S., Bussiere,D.E., Davies,C., White,S.W. and Bastia,D. (1996) Helicase–contrahelicase interaction and the mechanism of termination of DNA replication. Cell, 87, 881–891. [DOI] [PubMed] [Google Scholar]
- Marabini R., Masegosa,I.M., San Martín,M.C., Marco,S., Fernández,J.J., de la Fraga,L.G., Vaquerizo,C. and Carazo,J.M. (1996) Xmipp: an image processing package for electron microscopy. J. Struct. Biol., 116, 237–240. [DOI] [PubMed] [Google Scholar]
- Marabini R., Herman,G.T. and Carazo,J.M. (1998) 3D reconstruction in electron microscopy using ART with smooth spherically symmetric volume elements (blobs). Ultramicroscopy, 72, 53–65. [DOI] [PubMed] [Google Scholar]
- Miles C.S., Weigelt,J., Stamford,N.P.J., Dammerova,N., Otting,G. and Dixon,N.E. (1997) Precise limits of the N-terminal domain of DnaB helicase determined by NMR spectroscopy. Biochem. Biophys. Res. Commun., 231, 126–130. [DOI] [PubMed] [Google Scholar]
- Nakayama N., Arai,N., Kaziro,Y. and Arai,K. (1984) Structural and functional studies of the dnaB protein using limited proteolysis. Characterization of domains for DNA-dependent ATP hydrolysis and for protein association in the primosome. J. Biol. Chem., 259, 88–96. [PubMed] [Google Scholar]
- Patel S.S. and Picha,K.M. (2000) Structure and function of hexameric helicases. Annu. Rev. Biochem., 69, 651–697. [DOI] [PubMed] [Google Scholar]
- Radermacher M. (1994) Three-dimensional reconstruction from random projections: orientational alignment via Radon transforms. Ultramicroscopy, 53, 121–136. [DOI] [PubMed] [Google Scholar]
- Radermacher M. (1997) Radon transforms techniques for alignment and 3-D reconstruction from random projections. Scanning Microsc. Int. Suppl., 169–176. [Google Scholar]
- San Martín M.C., Stamford,N.P.J., Dammerova,N., Dixon,N.E. and Carazo,J.M. (1995) A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J. Struct. Biol., 114, 167–176. [DOI] [PubMed] [Google Scholar]
- San Martín M.C., Gruss,C. and Carazo,J.M. (1997) Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J. Mol. Biol., 268, 15–20. [DOI] [PubMed] [Google Scholar]
- San Martín M.C., Radermacher,M., Wolpensinger,B., Engel,A., Miles, C.S., Dixon,N.E. and Carazo,J.M. (1998) Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC. Structure, 6, 501–509. [DOI] [PubMed] [Google Scholar]
- Sawaya M.R., Guo,S., Tabor,S., Richardson,C.C. and Ellenberger,T. (1999) Crystal structure of the helicase domain from the replicative helicase–primase of bacteriophage T7. Cell, 99, 167–177. [DOI] [PubMed] [Google Scholar]
- Saxton W.O. and Baumeister,W. (1982) The correlation averaging of a regularly arranged bacterial cell envelope protein. J. Microsc., 127, 127–138. [DOI] [PubMed] [Google Scholar]
- Scherzinger E., Zieglin,G., Bárcena,M., Carazo,J.M., Lurz,R. and Lanka,E. (1997) The RepA protein of plasmid RSF1010 is a replicative DNA helicase. J. Biol. Chem., 272, 30228–30236. [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Sawaya,M.R., Ellenberger,T. and Wigley,D.B. (2000) Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell, 101, 589–600. [DOI] [PubMed] [Google Scholar]
- Squire P.G. and Himmel,M.E. (1979) Hydrodynamics and protein hydration. Arch. Biochem. Biophys., 196, 165–177. [DOI] [PubMed] [Google Scholar]
- Valle M., Gruss,C., Halmer,L., Carazo,J.M. and Donate,L.E. (2000) Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol. Cell. Biol., 20, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Lasken,R.S. and Kornberg,A. (1989a) The dnaB–dnaC replication protein complex of Escherichia coli. I. Formation and properties. J. Biol. Chem., 264, 2463–2468. [PubMed] [Google Scholar]
- Wahle E., Lasken,R.S. and Kornberg,A. (1989b) The dnaB–dnaC replication protein complex of Escherichia coli. II. Role of the complex in mobilizing dnaB functions. J. Biol. Chem., 264, 2469–2475. [PubMed] [Google Scholar]
- Weigelt J., Brown,S.E., Miles,C.S., Dixon,N.E. and Otting,G. (1999) NMR structure of the N-terminal domain of E.coli DnaB helicase: implications for structure rearrangements in the helicase hexamer. Structure, 7, 681–690. [DOI] [PubMed] [Google Scholar]
- Wickner S. and Hurwitz,J. (1975) Interaction of Escherichia coli dnaB and dnaC(D) gene products in vitro. Proc. Natl Acad. Sci. USA, 72, 921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Jezewska,M.J., Bujalowski,W. and Egelman,E.H. (1996) The hexameric E.coli DnaB helicase can exist in different quaternary states. J. Mol. Biol., 259, 7–14. [DOI] [PubMed] [Google Scholar]