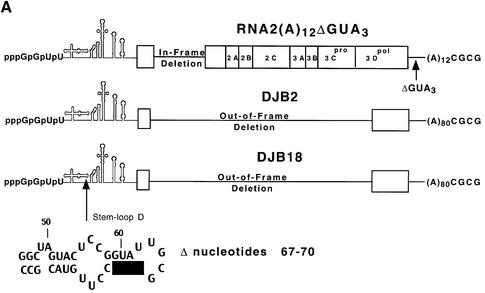
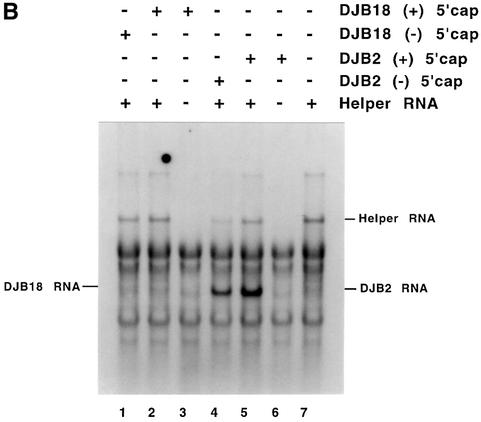
Fig. 4. Complementation analysis of DJB2 and DJB18 RNA replication in vitro. (A) Diagram of RNA2(A)12ΔGUA3, the helper RNA used in this experiment. The co-translation of this helper RNA provided the viral replication proteins in trans in reactions containing viral transcript RNAs that contained a deletion in the coding sequence for the viral replication proteins. DJB2 RNA does not encode any of the viral replication proteins but was shown in a separate study in our laboratory to contain all of the cis-active replication elements required to act as a template for negative-strand RNA synthesis. DJB18 RNA is identical to DJB2 RNA except for the deletion of poliovirus nucleotides 67–70 in the 5′ cloverleaf. (B) HeLa S10 translation–RNA replication reactions (100 µl) that contained 2 mM guanidine HCl and the indicated RNA (±) 5′ cap were incubated for 4 h at 34°C. The helper RNA was added at a 2:1 molar ratio relative to DJB2 RNA or DJB18 RNA, and the total RNA concentration was maintained at 100 µg/ml in each reaction. Negative-strand RNA synthesis was measured in preinitiation RNA replication complexes that were isolated from these reactions as described in Materials and methods. Reactions containing the preinitiation RNA replication complexes were incubated at 37°C for 25 min, and labeled negative-strand synthesis was analyzed by CH3HgOH–agarose gel electrophoresis. The mobility of DJB2 RNA, DJB18 RNA and the helper RNAs is shown. Control experiments showed that equivalent amounts of the viral replication proteins were synthesized in each reaction that contained the helper RNA (data not shown).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.