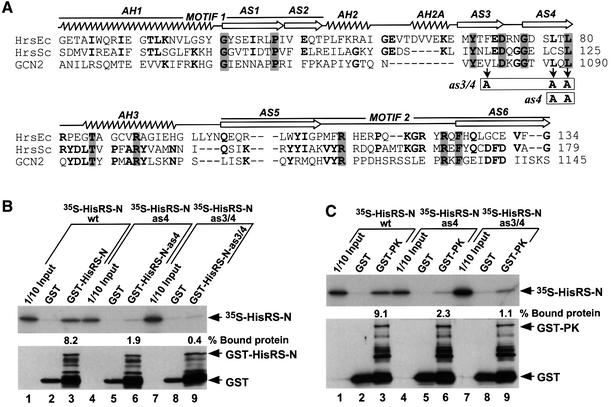
Fig. 4. Point mutations in the HisRS-N region abolish self-interaction and impair interaction with the PK domain in vitro. (A) Alignment of the HisRS-N region of GCN2 (amino acids 1036–1145) with segments from authentic histidyl-tRNA synthetases of E.coli (HrsEc) and Saccharomyces cerevisiae (HrsSc) encompassing motifs M1 and M2, the α-helices (AH1–AH3) and β-strands (AS1–AS6) present in the crystal structure of E.coli HisRS (Arnez et al., 1995), all indicated above the structure. The single or double mutations in β-strands AS3 (V1080A) and AS4 (L1088A, L1090A), designated as4 and as3/4, are indicated with boxes beneath the GCN2 sequence. (B and C) In vitro binding of [35S]HisRS-N segments (aa 970–1156) synthesized in vitro containing mutations as4 or as3/4 to GST fusion proteins containing the same HisRS-N segments (B), or the PK domain (C). The upper panels display the fluorograms of 35S-labeled proteins added to the reactions (1/10 Input) or bound to the GST or GST–HisRS-N proteins, as indicated. The percentages of the total 35S-labeled proteins bound in each reaction were quantified by phosphoimaging analysis and are indicated below the relevant lanes. The lower panels show the results of immunoblot analysis using GST antibodies of the GST proteins bound to glutathionine–Sepharose beads in each reaction. The positions of GST, GST–HisRS-N and GST–PK fusion proteins are indicated on the right.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.