TABLE 2.
Estimates of the average coefficient of dominance for segregating mutations and other parameters for a model of many mutations of low average effect
| Estimates of average h
|
% chromosomes
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean effect | by.x | R | 1/bx.y | hDL | QN | SL | D | VA | VD | VA/D |
| 0 (0, 0) | — | 0.07 | 0.15 | 0.28 | 0.09 | 88 | 12 | 0.169 | 0.0009 | 0.0005 | 0.005 |
| 50 (25, 25) | 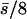 |
0.11 | 0.22 | 0.42 | 0.14 | 88 | 12 | 0.187 | 0.0017 | 0.0006 | 0.010 |
| 100 (50, 50) | 0.14 | 0.26 | 0.51 | 0.17 | 86 | 14 | 0.210 | 0.0025 | 0.0008 | 0.012 | |
| 150 (74, 76) | 0.18 | 0.29 | 0.56 | 0.20 | 85 | 15 | 0.233 | 0.0033 | 0.0009 | 0.014 | |
| 300 (149, 151) | 0.27 | 0.35 | 0.64 | 0.26 | 82 | 18 | 0.300 | 0.0057 | 0.0013 | 0.019 | |
| 10 (5, 5) | 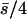 |
0.10 | 0.18 | 0.39 | 0.13 | 88 | 12 | 0.179 | 0.0021 | 0.0011 | 0.012 |
| 50 (23, 27) | 0.21 | 0.27 | 0.58 | 0.23 | 85 | 15 | 0.226 | 0.0051 | 0.0017 | 0.023 | |
| 100 (47, 53) | 0.29 | 0.32 | 0.67 | 0.30 | 81 | 19 | 0.283 | 0.0092 | 0.0025 | 0.032 | |
| 150 (70, 80) | 0.34 | 0.35 | 0.71 | 0.34 | 75 | 25 | 0.332 | 0.0129 | 0.0032 | 0.039 | |
| 10 (4, 6) |
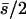
|
0.16 | 0.21 | 0.51 | 0.20 | 87 | 13 | 0.190 | 0.0046 | 0.0019 | 0.024 |
| 50 (21, 29) | 0.32 | 0.32 | 0.78 | 0.38 | 73 | 27 | 0.318 | 0.0182 | 0.0061 | 0.057 | |
| 100 (42, 58) | 0.35 | 0.37 | 0.95 | 0.45 | 51 | 49 | 0.486 | 0.0384 | 0.0115 | 0.079 | |
| 150 (63, 87) | 0.35 | 0.40 | 1.21 | 0.48 | 34 | 66 | 0.647 | 0.0552 | 0.0168 | 0.085 | |
Nonlethal mutational parameters: λ = 0.04, 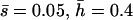 .
.
Other parameters and definitions are as in Table 1. The number of segregating nonpleiotropic mutations in the chromosome sample was 2396 ± 45. Ranges of SE: by.x, 0.002–0.005; R, 0.001–0.004; 1/bx.y, 0.003–0.028; hDL, 0.003–0.008; QN, 0.59–1.35; SL, 0.76–1.40; D, 0.005–0.010; VA, 0.0000–0.0006; and VD, 0.0000–0.0003.
