TABLE 3.
Estimates of the average coefficient of dominance for segregating mutations and other population parameters for a model where nonantagonistic mutations show strengthening pleiotropy
| Estimates of average h
|
% chromosomes
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| n | by.x | R | 1/bx.y | hDL | QN | SL | D | VA | VD | VA/D |
| A. | ||||||||||
| 0 (0, 0) | 0.04 | 0.03 | 0.17 | 0.05 | 92 | 8 | 0.069 | 0.0006 | 0.0006 | 0.009 |
| 3 (1, 2) | 0.23 | 0.17 | 0.58 | 0.21 | 90 | 10 | 0.109 | 0.0082 | 0.0033 | 0.075 |
| 5 (1, 4) | 0.32 | 0.23 | 0.68 | 0.30 | 87 | 13 | 0.151 | 0.0139 | 0.0053 | 0.092 |
| 10 (2, 8) | 0.38 | 0.30 | 0.80 | 0.41 | 77 | 23 | 0.249 | 0.0271 | 0.0102 | 0.109 |
| 15 (3, 12) | 0.37 | 0.33 | 0.91 | 0.46 | 65 | 35 | 0.349 | 0.0401 | 0.0147 | 0.115 |
| B. | ||||||||||
| 0 (0, 0) | 0.06 | 0.13 | 0.25 | 0.08 | 93 | 7 | 0.100 | 0.0016 | 0.0005 | 0.016 |
| 10 (4, 6) | 0.19 | 0.21 | 0.51 | 0.22 | 92 | 8 | 0.130 | 0.0053 | 0.0016 | 0.041 |
| 50 (21, 29) | 0.36 | 0.33 | 0.74 | 0.42 | 80 | 20 | 0.260 | 0.0195 | 0.0058 | 0.075 |
| 100 (42, 58) | 0.37 | 0.38 | 0.90 | 0.48 | 60 | 40 | 0.416 | 0.0364 | 0.0107 | 0.087 |
| 150 (63, 87) | 0.37 | 0.41 | 1.08 | 0.50 | 40 | 60 | 0.585 | 0.0554 | 0.0162 | 0.095 |
Nonlethal mutational parameters: (A) λ = 0.006, 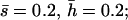 (B) λ = 0.04,
(B) λ = 0.04, 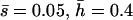 . The number of segregating strengthening pleiotropic mutations in the chromosome sample was 70 ± 1 for model A and 2102 ± 22 for model B. The mean selection coefficient of antagonistic pleiotropic mutations is
. The number of segregating strengthening pleiotropic mutations in the chromosome sample was 70 ± 1 for model A and 2102 ± 22 for model B. The mean selection coefficient of antagonistic pleiotropic mutations is 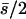 . Other parameters and definitions are as in Table 1. Ranges of SE: by.x, 0.001–0.005; R, 0.001–0.005; 1/bx.y, 0.002–0.011; hDL, 0.001–0.008; QN, 0.26–0.79; SL, 0.31–0.69; D, 0.002–0.006; VA 0.0000–0.0010; and VD, 0.0000–0.0004.
. Other parameters and definitions are as in Table 1. Ranges of SE: by.x, 0.001–0.005; R, 0.001–0.005; 1/bx.y, 0.002–0.011; hDL, 0.001–0.008; QN, 0.26–0.79; SL, 0.31–0.69; D, 0.002–0.006; VA 0.0000–0.0010; and VD, 0.0000–0.0004.
