TABLE 4.
Estimates of the average coefficient of dominance for segregating mutations and other parameters for a model of few mutations of large average effect, under different population sizes (A) and an asymmetric model of pleiotropy (B)
| Estimates of average h
|
% chromosomes
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | n | by.x | R | 1/bx.y | hDL | QN | SL | D | VA | VD | VA/D |
| A. | |||||||||||
| 102 | 0 | 0.15 | 0.15 | 0.33 | 0.18 | 98 | 2 | 0.018 | 0.0003 | 0.0005 | 0.017 |
| 15 | 0.39 | 0.34 | 0.62 | 0.44 | 96 | 4 | 0.095 | 0.0074 | 0.0023 | 0.078 | |
| 103 | 0 | 0.07 | 0.07 | 0.25 | 0.10 | 94 | 6 | 0.056 | 0.0003 | 0.0007 | 0.005 |
| 15 | 0.35 | 0.31 | 0.65 | 0.37 | 91 | 9 | 0.150 | 0.0090 | 0.0029 | 0.060 | |
| 105 | 0 | 0.03 | 0.02 | 0.16 | 0.03 | 77 | 23 | 0.206 | 0.0002 | 0.0013 | 0.001 |
| 15 | 0.27 | 0.22 | 0.72 | 0.20 | 73 | 27 | 0.296 | 0.0089 | 0.0034 | 0.030 | |
| B. | |||||||||||
| 3 | 0.08 | 0.15 | 0.40 | 0.10 | 86 | 14 | 0.132 | 0.0014 | 0.0010 | 0.011 | |
| 104 | 5 | 0.11 | 0.22 | 0.51 | 0.13 | 86 | 14 | 0.139 | 0.0021 | 0.0012 | 0.015 |
| 10 | 0.18 | 0.31 | 0.62 | 0.19 | 85 | 15 | 0.157 | 0.0041 | 0.0015 | 0.026 | |
| 15 | 0.24 | 0.35 | 0.69 | 0.24 | 83 | 17 | 0.178 | 0.0060 | 0.0018 | 0.034 | |
Nonlethal mutational parameters: λ = 0.006, 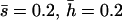 . N, population size. n, number of antagonistic pleiotropic mutations segregating per chromosome. The mean selection coefficient of antagonistic pleiotropic mutations is
. N, population size. n, number of antagonistic pleiotropic mutations segregating per chromosome. The mean selection coefficient of antagonistic pleiotropic mutations is 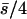 . Ranges of SE: by.x, 0.001–0.004; R, 0.000–0.003; 1/bx.y, 0.002–0.008; hDL, 0.001–0.008; QN, 0.18–0.81; SL, 0.11–0.39; D, 0.001–0.009; VA, 0.0000–0.0002; and VD, 0.0000–0.0001. Other parameters and definitions are as in Table 1.
. Ranges of SE: by.x, 0.001–0.004; R, 0.000–0.003; 1/bx.y, 0.002–0.008; hDL, 0.001–0.008; QN, 0.18–0.81; SL, 0.11–0.39; D, 0.001–0.009; VA, 0.0000–0.0002; and VD, 0.0000–0.0001. Other parameters and definitions are as in Table 1.
