Abstract
Several models have been proposed to explain why expression parameters of a gene might be related to the size of the gene's introns. These include the idea that an energetic cost of transcription should favor smaller introns in highly expressed genes (the “economy selection” argument) and that tissue-specific genes reside in genomic locations with complex chromatin level control requiring large amounts of noncoding DNA (the “genomic design” hypothesis). We recently proposed a modification of the economy model arguing that, for some genes, the time that expression takes is more important than the energetic cost, such that some weakly but rapidly expressed genes might also have small introns. We suggested that antisense genes might be such a class and showed that the data appear to be consistent with this. We now reexamine this model to ask (a) whether the effects described were owing solely to the fact that antisense genes are often noncoding RNA and (b) whether we can confidently reject the “genomic design” model as an explanation for the facts. We show that the effects are not specific to noncoding RNAs and that the predictions of the “genomic design” model for the most part are not upheld.
SEVERAL different models have been proposed to explain why broadly/highly expressed genes typically have small introns. The “economy selection” model argues that this phenomenon reflects selection for minimizing the costs of gene expression (Hurst et al. 1996; Castillo-Davis et al. 2002; Eisenberg and Levanon 2003; Seoighe et al. 2005), the “mutational bias” model suggests that it reflects regional mutation biases in rates of insertion and deletion (Urrutia and Hurst 2003), while the “genomic design” model postulates that it reflects selection for genomic organization to enable control of gene expression (Vinogradov 2004, 2005). Recently, we observed that human antisense genes have significantly shorter introns compared with other genes, including their more broadly/highly expressed sense partners. This, we argued, could not be simply explained by any of the above models. Our further analyses suggested that the short introns of antisense genes might be related to antisense regulation that requires a rapid response time (Chen et al. 2004, 2005a). Thus, we proposed an “efficiency selection” model (which can also be recognized as a “time-economy selection” model) to explain the short introns of antisense genes (Chen et al. 2005a). Here, in this short note, we address two matters. First, is the reduced intronic size of antisense genes true for protein-coding antisense as well as for noncoding RNA (ncRNA) or was our observation more a statement about ncRNA than about antisense per se? Note that the antisense genes are enriched for ncRNA: the percentage of the genes with an assigned protein-coding region (CDS) in the antisense (A), sense (S), antisense-like (AL), sense-like (SL), and nonoverlapping bidirection (NBD) genes (see their classification in the legend of Figure 1; see also Chen et al. 2004, 2005a,b) is 57.5, 94.5, 40.2, 91.6, and 79.1%, respectively. Second, have we been premature in our rejection of the genomic design model?
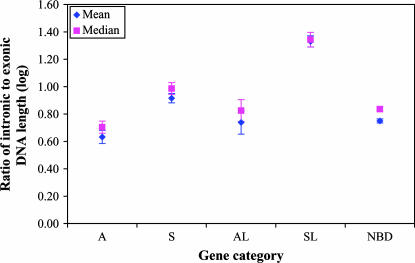
Figure 1.
Comparison of the ratios of intronic-to-exonic length among the five gene categories. The S (sense) and A (antisense) genes form SA (sense-antisense) gene pairs with exon overlaps, while the SL (sense-like) and AL (antisense-like) genes form NOB (non-exon-overlapping bidirectional) gene pairs without exon overlaps; both SA and NOB pairs are bidirectional (BD) gene pairs. NBD (nonbidirectional) genes contain only single-direction transcribed sequences. We classified the S and A or SL and AL genes in each bidirectional gene pair mainly on the basis of the conventional concept (e.g., Lipman 1997) that the S (or SL) gene should exist in more tissues and/or be expressed at a higher level than its A (or AL) partner gene (Chen et al. 2004, 2005a,b). The mean and median values with their 95% confidence intervals of the logarithm (log) values of the ratios of intronic-to-exonic DNA length are shown in the plot. Note that the original values are not normally distributed, but the logarithm values are almost normally distributed. Thus, we use independent samples t-test and Mann-Whitney U-test to determine significance (P-value) in their mean-value and median-value differences in the logarithm ratio data, respectively. The A and AL genes have significantly lower ratios of intronic-to-exonic DNA length (P < 10−4) compared with the S and SL genes, respectively. In addition, the mean and median values of the logarithm ratios in the AL genes are significantly lower (P < 0.001) than in the S genes and similar to the NBD genes. The A genes have the lowest ratios among the five gene categories, which is in accord with the efficiency selection model (Chen et al. 2005a). The detailed values of the original and log-transformed data as well as the significance (P-values) of the comparisons based on the logarithm values are shown in Table 1. In fact, we observed a similar pattern in analysis of the original data. Although the mean value of the original ratios is higher in the AL genes than in the S and NBD genes, the median value of the original ratios as well as the mean and median values of the logarithm ratios in the AL genes is significantly lower (P < 0.001) than in the S genes and similar to the NBD genes. Indeed, we observed that the high mean value of the original ratios in the AL genes was caused by a very small group of extreme big ratios (data not shown).
Small introns for protein-coding antisense genes:
To address the first issue, we excluded all the genes without CDS. However, this leaves a problem, namely which gene in a pair of bidirectional protein-coding genes should we consider the sense gene and which the antisense? We defined the S and A (or SL and AL) on the basis of the conventional concept (e.g., Lipman 1997) that the sense (or SL) gene should exist in more tissues and be expressed at a higher level than its antisense partner (Chen et al. 2004, 2005a,b). If the expression levels of the two paired genes are the same, the gene pair is excluded from this analysis. We should expect that if the efficiency selection model is wrong and the economy model is uniquely true, then the sense gene with the higher expression level should be the one with the smaller introns. If the efficiency selection model is correct, it could, in principle, be reversed. Therefore, to ask if the efficiency selection model might have merits, we consider whether antisense has smaller introns compared to its sense partner. If the more weakly expressed antisense gene has the smaller introns, this could not be explained by economy or regional mutational bias, but would be consistent with efficiency.
In analysis of the protein-coding genes only, we observed a pattern similar to the previous observation in which both protein-coding and noncoding genes are considered (Chen et al. 2005a), namely a reduced intronic size for the putative antisense genes: the average intron length of the protein-coding A, S, AL, SL, and NBD genes is 4779 nucleotides (nt) (average logarithm value: 3.3113), 5032 nt (3.3935), 9278 nt (3.4570), 12,137 nt (3.7340), and 4904 nt (3.3588), respectively, while the average intron lengths of the whole-gene sets are 4995 nt (3.3018), 5313 nt (3.3759), 9049 nt (3.4565), 12,599 nt (3.7338), and 5202 nt (3.3558), respectively. The differences are still significant between antisense genes and any other kind of genes (P < 0.01; both independent samples t-test and Mann-Whitney U-test were used in analyses of both the original and logarithim values), as well as between antisense genes and their sense partners (or between AL and SL genes) in a paired fashion (P < 0.05; paired samples t-test). Thus, the difference that we see between S (SL) and A (AL) genes does not result exclusively from the potential difference in intron size between protein-coding genes and ncRNA genes.
Predictions of genomic design for the most part are not upheld:
The genomic design hypothesis (Vinogradov 2004) proposed that lowly expressed genes have large introns as these introns may contain large suppressing control elements. The model also proposes that the noncoding DNA in and around a gene (gene nests) affects the expression of the gene through chromatin level regulation. It is this feature that predicts that a gene with small introns should sit in a region with small intergenic distance and should have genes of intronic dimensions comparable to its neighbors. The model predicts, we reasoned, that in a comparison of pairs of linked genes, the one with lower/narrower expression should be the one with the larger introns, as it should be the one with the extra control elements. This is the opposite of what we found in comparing antisense genes with their overlapping sense partners in a paired fashion (Chen et al. 2005a), namely that antisense genes have significantly shorter introns (P < 0.01) than their sense partners, although their sense partners have significantly higher expression levels and wider expression breadths (P < 10−4). We therefore rejected this as a potential explanation for our set of observed facts. The same finding rejects biased mutation and economy selection, the former predicting no difference between the genes in mean intron size of neighbors, the latter predicting smaller introns for the more highly expressed gene (see also supplemental Figure 1, a and b, at http://www.genetics.org/supplemental/).
The genomic design model makes further predictions (A. E. Vinogradov, personal communication). It suggests that, although intronic sequences of the A and AL genes are shorter than those of the S and SL genes, respectively, this may be consistent with the genomic design model if we were to find that genes residing in the same genomic region have a similar intronic-to-exonic length ratio (because chromatin of the gene nest condenses and decondenses as a whole).
To test the prediction, we compared the intronic and exonic DNA lengths as well as the ratio of intronic-to-exonic length in the A, S, AL, SL, and NBD genes. In the following analyses, we did not exclude noncoding genes to be consistent with our previous study (Chen et al. 2005a). Because the full lengths for both intronic and exonic sequences are necessary for the analysis, we focus on full-length mRNA sequences that also span introns (although we observed a similar pattern in analysis of all intron-spanning sequences; data not shown). Our findings do not support the prediction. Although the A and AL genes are shorter (compared to the S and SL genes) in both intron and exon (supplemental Figure 2 at http://www.genetics.org/supplemental/), on average the A and AL genes have a significantly lower ratio of intronic-to-exonic length (P < 10−4) compared to the S and SL genes, respectively (Figure 1, Table 1). This is also true in a comparison of the A genes with their S partners and the AL genes with their SL partners when considered in a paired fashion. We have collected 397 S/A and 205 SL/AL gene pairs in which both members have full-length, intron-spanning mRNA sequences. The A and AL genes have a significantly lower ratio of intronic-to-exonic length (P < 10−4; paired samples t-test with the logarithm ratios) than do their S and SL partners (data not shown). Thus, contrary to the genomic design hypothesis, we find no evidence that the intron/exon ratio for sense and antisense genes is a property of the genomic region (gene nest) within which these genes reside.
TABLE 1.
Comparisons of ratios and P-values of the five gene categories
| Ratio of intronic-to-exonic DNA length
|
||||
|---|---|---|---|---|
| Gene category | Mean of the original ratios | Median of the original ratios | Mean of the log ratios | Median of the log ratios |
| A (n = 855) | 13.24 | 5.06 | 0.63 | 0.70 |
| S (n = 1318) | 19.05 | 9.68 | 0.92 | 0.99 |
| AL (n = 390) | 24.22 | 6.78 | 0.74 | 0.83 |
| SL (n = 769) | 45.65 | 22.03 | 1.33 | 1.34 |
| NBD (n = 7942) | 17.15 | 6.85 | 0.75 | 0.84 |
| Significance (i.e., P-value) of difference in the ratios of intronic-to-exonic length determined by t-test and U-test based on the log ratios
| |||||
|---|---|---|---|---|---|
| Gene category | A | S | AL | SL | NBD |
| A | — | <10−4 (<10−4) | <0.01 (<0.01) | <10−4 (<10−4) | <10−4 (<10−4) |
| S | — | — | <10−4 (<0.001) | <10−4 (<10−4) | <10−4 (<10−4) |
| AL | — | — | — | <10−4 (<10−4) | >0.1 (>0.1) |
| SL | — | — | — | — | <10−4 (<10−4) |
| NBD | — | — | — | — | — |
A. E. Vinogradov (personal communication) also predicts that the intronic-to-exonic length ratio should be higher in the SL/AL genes (compared to the S/A genes and nonoverlapping genes) owing to the exon-intron overlap of the opposite genes in this pair (i.e., the intronic sequences of these genes do not consist completely of noncoding DNA because they may encode exons on the opposite strand). The evidence here is mixed. Contrary to the prediction, on average the ratio of intronic-to-exonic length of the AL genes is significantly lower than (P < 10−2) not only that of the SL genes but also that of the S genes and similar to that of the NBD genes (Figure 1). However, in agreement with the prediction, the SL genes do have the highest ratio of all classes and the AL genes have a higher ratio than the A genes (Figure 1). The balance of evidence, however, suggests that the genomic design model cannot provide the complete explanation. Notably, the fact that AL genes have a lower ratio than SL genes is important because, if SL genes have large introns owing to inclusion of exons of the antisense within the SL introns, by the same logic AL genes should have still larger introns as SL exons are larger than AL exons (supplemental Figure 2 at http://www.genetics.org/supplemental/). That the reverse is found argues against the genomic design model.
While the observations for the most part appear to be inconsistent with the genomic design hypothesis, how might they be explained? Economy selection (of energy or of time) can be realized in one of the counterpart genes of a given pair and its effect should be stronger on the intronic than on the exonic part of the gene (because the latter is under stronger functional constraints). As a result, the ratios of intronic-to-exonic length would differ between counterpart genes, just as we observed (Figure 1). As the A and AL genes have a significantly lower and narrower expression compared with the S and SL genes (supplemental Figure 1, a and b, at http://www.genetics.org/supplemental/), the “energy-economy selection” model cannot explain the observations. Thus, only the “time-economy selection” (i.e., the efficiency selection) model (Chen et al. 2005a) can provide a feasible explanation. This notion is further bolstered by the finding that antisense genes in the coexpressed, inversely expressed, and/or evolutionarily conserved SA pairs (these classes of SA pairs being the most likely to participate in antisense-mediated gene regulation that requires a rapid response time; Chen et al. 2005b) have the most extremely short introns (Chen et al. 2005a). Given this, it is not surprising that the antisense genes have a significantly lower ratio of intronic-to-exonic length than do the S genes (Figure 1).
Why do antisense-like genes have small introns?
One issue that we did not consider previously was whether the efficiency selection model might additionally explain the significant difference between the AL and SL genes in the ratio of intronic-to-exonic length. Although the regulatory interactions were presumed only for the SA pairs as they have exon overlaps (Knee and Murphy 1997; Kumar and Carmichael 1998; Vanhee-Brossollet and Vaquero 1998; Chen et al. 2004, 2005a,b), theoretically, it is possible for an AL gene to regulate the expression of its SL partner if their pre-mRNA molecules could form double-stranded RNAs in the nucleus. Since double-stranded secondary structures formed by base pairing between exons and downstream intron elements in pre-mRNAs of the same genes (i.e., exon-intron duplex structures) have been observed in many cases (see Wang and Carmichael 2004 and references therein), it is reasonable to presume that double-stranded RNAs can be formed by base pairing between exons (and/or introns) and cognate introns (i.e., forming exon-intron and/or intron-intron duplexes) of the counterpart genes in a given SL/AL gene pair before the overlapped intronic regions in pre-mRNAs are spliced out. Indeed, the fact that, as with SA pairs, human SL/AL pairs are also significantly more frequently (P < 0.05) coexpressed than are pseudo-gene-pair sets with the same expression levels (Chen et al. 2005b, supplemental Table 3) provides indirect evidence to support the hypothesis that regulation may also exist between AL and SL. Nevertheless, there is no direct experimental evidence yet; thus, a systematic and genome-wide identification of all naturally occurring double-stranded RNAs in the nucleus of some cell types would be necessary to test the hypothesis.
Conclusion:
In sum, predictions of the genomic design model regarding the ratios of intronic-to-exonic length between antisense (antisense-like) genes and sense (sense-like) genes for the most part are not upheld and hence the above results argue against this model as a means to account for the unusually small introns of antisense genes. Both protein-coding and noncoding antisense genes show the same reduced intronic dimensions. More generally, the results strengthen our previous conclusion (Chen et al. 2005a) that the findings are inconsistent with prior models but are consistent with efficiency selection. We should, however, reiterate that we do not propose that the efficiency selection model explains all variation in intronic sizes. We still consider the genomic design model as one of several models that need to be considered in the more general context of understanding intergene variation in intronic dimensions, not least because it is one of the few models that attempts to account for the correlation between intron size and intergenic distance (Vinogradov 2004, 2005). Indeed, it is worth noting that the SL genes have much longer introns (Chen et al. 2005a; supplemental Figure 2 at http://www.genetics.org/supplemental/) and a much higher ratio of intronic-to-exonic length (Figure 1) than do the NBD genes although the SL genes have significantly higher expression levels and wider expression breadths than do the NBD genes (supplemental Figure 1 at http://www.genetics.org/supplemental/). This observation could not be explained by either our model or the economy selection model, but can be explained by the genomic design model. The genomic design model proposed that the phenomenon that SL genes have much longer introns than do NBD genes is due to the fact that the intronic sequences of the SL genes do not completely consist of noncoding DNA because they may encode exons on the opposite strand for the AL genes (A. E. Vinogradov, personal communication).
Acknowledgments
We thank Alexander E. Vinogradov for allowing us to cite his thoughtful prediction and hypothesis as personal communication and Laurent Duret for his constructive discussions. This work was supported by the G. Harold and Leila Y. Mathers Charitable Foundation (J.C.), National Institutes of Health grant CA84405 (J.D.R.), the Spastic Paralysis Foundation of the Illinois, Eastern Iowa Branch of Kiwanis International (J.D.R.), and the U.K. Biotechnology and Biological Sciences Research Council (L.D.H.).
References
- Castillo-Davis, C. I., S. L. Mekhedov, D. L. Hartl, E. V. Koonin and F. A. Kondrashov, 2002. Selection for short introns in highly expressed genes. Nat. Genet. 31: 415–418. [DOI] [PubMed] [Google Scholar]
- Chen, J., M. Sun, W. J. Kent, X. Huang, H. Xie et al., 2004. Over 20% of human transcripts might form sense–antisense pairs. Nucleic Acids Res. 32: 4812–4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., M. Sun, L. D. Hurst, G. G. Carmichael and J. D. Rowley, 2005. a Human antisense genes have unusually short introns: evidence for selection for rapid transcription. Trends Genet. 21: 203–207. [DOI] [PubMed] [Google Scholar]
- Chen, J., M. Sun, L. D. Hurst, G. G. Carmichael and J. D. Rowley, 2005. b Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 21: 326–329. [DOI] [PubMed] [Google Scholar]
- Eisenberg, E., and E. Y. Levanon, 2003. Human housekeeping genes are compact. Trends Genet. 19: 362–365. [DOI] [PubMed] [Google Scholar]
- Hurst, L. D., G. McVean and T. Moore, 1996. Imprinted genes have few and small introns. Nat. Genet. 12: 234–237. [DOI] [PubMed] [Google Scholar]
- Knee, R., and P. R. Murphy, 1997. Regulation of gene expression by natural antisense RNA transcripts. Neurochem. Int. 31: 379–392. [DOI] [PubMed] [Google Scholar]
- Kumar, M., and G. G. Carmichael, 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62: 1415–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman, D. J., 1997. Making (anti)sense of non-coding sequence conservation. Nucleic Acids Res. 25: 3580–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoighe, C., C. Gehring and L. D. Hurst, 2005. Gametophytic selection in Arabidopsis thaliana supports the selective model of intron length reduction. PLoS Genet. 1: e13. [DOI] [PMC free article] [PubMed]
- Urrutia, A. O., and L. D. Hurst, 2003. The signature of selection mediated by expression on human genes. Genome Res. 13: 2260–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhee-Brossollet, C., and C. Vaquero, 1998. Do natural antisense transcripts make sense in eukaryotes? Gene 211: 1–9. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A. E., 2004. Compactness of human housekeeping genes: Selection for economy or genomic design? Trends Genet. 20: 248–253. [DOI] [PubMed] [Google Scholar]
- Vinogradov, A. E., 2005. Noncoding DNA, isochores and gene expression: nucleosome formation potential. Nucleic Acids Res. 33: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., and G. G. Carmichael, 2004. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 68: 432–452. [DOI] [PMC free article] [PubMed] [Google Scholar]