Figure 1.
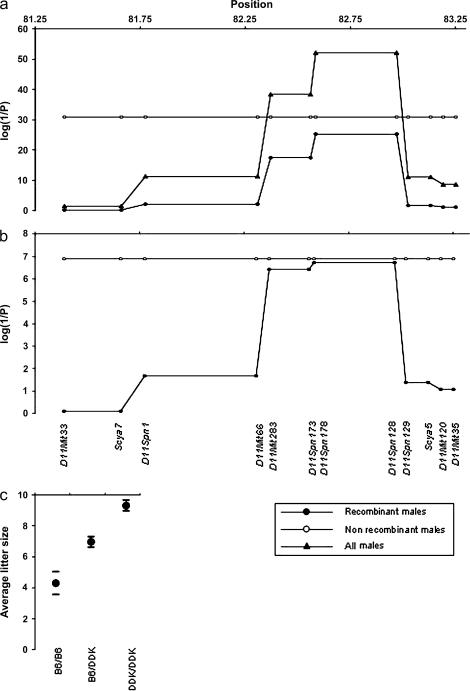
Mapping by recombinant progeny testing in C57BL/6–DDK background. The reproductive performances of 40 males were used to map the paternal gene in the candidate interval defined previously (Baldacci et al.1992, 1996). The original interval was divided into six smaller intervals based on the presence of five recombinant chromosomes. The markers shown in the bottom of section b convey the complete genotypic information (supplemental Table 2 at http://www.genetics.org/supplemental/). In sections a and b the horizontal axes show distance in Mbp from the centromere of chromosome 11. The vertical axes show the statistical significance of the association findings. Triangles show the results using the entire set of 40 males. Open circles show the results for the 22 males carrying nonrecombinant chromosomes in the D11Mit33–D11Mit35 interval. Filled circles show the results in the 18 males carrying recombinant chromosomes in the D11Mit3–D11Mit35 interval (the inset in the bottom right shows this information visually). (a) The top plot shows the strength of the associations between genotype and litter size using the Fdistribution to assess the statistical significance of the mixed models. (b) The bottom plot shows the strength of the association between genotype and litter size using permutation tests to assess the statistical significance of the mixed models. Empirical results for the 22 nonrecombinant males and for the regions containing markers D11Spn173 and D11Spn178 among the 18 recombinant males were determined exactly. The statistical significance when analyzing all 40 males for the regions containing marker D11Mit33 (P = 0.045) was estimated using a permutation test while the statistical significance for D11Spn173 (P = 1.3 × 10−15) and D11Spn178 (P = 3.4 × 10−16) were calculated exactly (data not shown). It was not feasible to calculate the empirical statistical significance for the other regions (P < 1.0 × 10−5) using all 40 males. The statistical significance of the findings in these other regions were beyond the resolution of 100,000 random replications of the data and there were too many permutations of the data that would yield a more extreme value than the observed F-statistics to make it possible to determine the empirical significance exactly. (c) Circles represent the average litter size in males with the three possible genotypes in the D11Spn178–D11Spn128. The horizontal bars show the boundaries of the 99% confidence intervals.