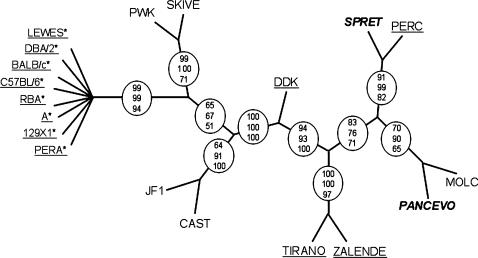
Figure 6.
Phylogenetic relationships of inbred strains in the 128 kb interval defined by in silico mapping. The tree depicted in the figure is a consensus cladogram from three consensus trees obtained by three different phylogenetic methods (materials and methods). Circles denote branches that are consistent among the three trees and the numbers in the circles represent the number of times out of 100 that the branch is observed in each method: top, DNAML; middle, NEIGHBOR; and bottom, DNAPARS. Underlined strains have M. m. domesticus haplotypes in that region. Strains in boldface and italics are derived from other species (SPRET, M. spretus and PANCEVO, M. spicilegus). Asterisks denote strains with incompatible alleles at the paternal gene. All incompatible strains are shown diverging from a single node because the internal branching order within this lineage is not consistent among trees obtained by different methods and the branching order is poorly supported within each method. The length of the branches is arbitrary.