TABLE 3.
Type I error rates (percentage) of test statistics FAB,ad, FAB,a, and FHTR of the haplotype trend regression (HTR) method at a 0.05 significance level when two markers A and B are used in the analysis
LD measure 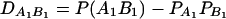
|
Error Rates
|
||||
|---|---|---|---|---|---|
| Sample size | Test case | FAB,ad | FAB,a | FHTR | |
| 0 | N = 200 | Null | 4.90 | 5.22 | 5.39 |
| Additive | 5.09 | 4.75 | 4.77 | ||
| Dominant | 4.62 | 4.87 | 4.79 | ||
| Recessive | 5.36 | 5.12 | 4.81 | ||
| 0.08 | N = 200 | Null | 5.09 | 5.23 | 5.55 |
| Additive | 4.92 | 4.74 | 4.71 | ||
| Dominant | 4.63 | 4.84 | 4.71 | ||
| Recessive | 5.04 | 5.02 | 4.94 | ||
The total variance is fixed as σ2 = 1.0 and the trait allele frequency is taken as q1 = q2 = 0.5. The numbers m and n of alleles = 2. The allele frequencies are given by 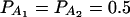 and
and 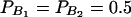 . Four test cases are considered: null, no major gene effect a = d = 0; additive, additive mode of inheritance a = 1, but no dominant effect d = 0; dominant, dominant mode of inheritance a = d = 1; recessive, recessive mode of inheritance a = 1 and d = –0.5. In each test case, linkage equilibrium is assumed between the QTL Q and the markers A and B; i.e.,
. Four test cases are considered: null, no major gene effect a = d = 0; additive, additive mode of inheritance a = 1, but no dominant effect d = 0; dominant, dominant mode of inheritance a = d = 1; recessive, recessive mode of inheritance a = 1 and d = –0.5. In each test case, linkage equilibrium is assumed between the QTL Q and the markers A and B; i.e., 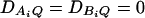 . No linkage disequilibrium of third order is assumed among markers A and B and the QTL Q; that is, DAQB = 0.
. No linkage disequilibrium of third order is assumed among markers A and B and the QTL Q; that is, DAQB = 0.
