Abstract
Fluoxetine (Prozac) is one of the most widely prescribed pharmaceuticals, yet important aspects of its mechanism of action remain unknown. We previously reported that fluoxetine and related antidepressants induce nose muscle contraction of C. elegans. We also reported the identification and initial characterization of mutations in seven C. elegans genes that cause defects in this response (Nrf, nose resistant to fluoxetine). Here we present genetic evidence that the known nrf genes can be divided into two subgroups that confer sensitivity to fluoxetine-induced nose contraction by distinct pathways. Using both tissue-specific promoters and genetic mosaic analysis, we show that a gene from one of these classes, nrf-6, functions in the intestine to confer fluoxetine sensitivity. Finally, we molecularly identify nrf-5, another gene in the same class. The NRF-5 protein is homologous to a family of secreted lipid-binding proteins with broad ligand specificity. NRF-5 is expressed in the intestine and is likely secreted into the pseudocoelomic fluid, where it could function to transport fluoxetine. One model that explains these findings is that NRF-5 binds fluoxetine and influences its presentation or availability to in vivo targets.
UNDERSTANDING the molecular mechanisms of the action of drugs is essential for rational development and delivery of compounds with greater efficacy, greater specificity, and fewer adverse side effects. Genetic screens for mutants with altered responses to drugs constitute an approach to identifying drug mechanisms of action that is particularly promising for identifying novel targets and pathways (Schafer 1999). Genetic screens using Caenorhabditis elegans have been successful in identifying drug targets. For example, screens for resistance to the antihelminthic drug levamisole led to the identification of levamisole-sensitive acetylcholine receptors (Lewis et al. 1980; Fleming et al. 1997) and screens for resistance to another antihelminthic drug, ivermectin, led to the identification of a novel class of ivermectin-sensitive glutamate-gated chloride channels (Dent et al. 2000). In addition to identifying direct drug targets, genetic resistance screens can also identify components of signaling pathways that are affected by the drug of interest. For example, the acetylcholinesterase inhibitor aldicarb causes increased synaptic acetylcholine levels and aldicarb-resistant mutants have identified several novel components of synaptic transmission (Nguyen et al. 1995; Miller et al. 1996, 2000) Finally, such screens can identify genes involved in drug metabolism, transport, or localization, which are key factors in determining in vivo drug availability and efficacy.
We isolated mutations affecting seven Nrf genes that cause resistance to nose contraction by the antidepressant fluoxetine (Nrf: nose resistant to fluoxetine). Genetic analysis suggested that three of these genes, nrf-5, nrf-6, and ndg-4, function in a common pathway and that mutations in these genes confer their fluoxetine-resistant phenotype by a similar mechanism (Choy and Thomas 1999). Here, we describe further genetic and molecular characterization of the Nrf genes. We construct double mutants between two classes of Nrf mutants and demonstrate that these two classes confer fluoxetine resistance by different pathways. We have examined the site of action of nrf-6 and found that it is required in the intestine for sensitivity to fluoxetine-induced nose contraction. We have also cloned nrf-5 and found that it is homologous to a family of mammalian secreted lipid-binding proteins. A nrf-5∷gfp fusion is expressed in the intestine, suggesting that nrf-5 is secreted into the pseudocoelomic fluid where it could function in drug transport.
MATERIALS AND METHODS
Genetics and pharmacology:
General culturing and maintenance of strains was as described (Brenner 1974). For nrf (Peg: pale eggs); nrf (non-Peg) double mutants, the Peg mutation was followed by the Peg phenotype and the non-Peg mutation was balanced in trans with flanking double-mutant chromosomes (Choy and Thomas 1999). To construct nrf-2; nrf-3 double mutants, both mutations were balanced in trans. All double mutants were confirmed by complementation testing. Nrf assays were performed as previously described (Choy and Thomas 1999).
Mosaic analysis:
nrf-6(sa367); lin-15(n756ts) mutants were injected with the nrf-6-rescuing cosmid C08B11 (5 ng/μl), pbLH98 lin-15(+) (60 ng/μl) (Clark et al. 1994), and pTJ1286 sur-5∷gfp (100 ng/μl) (Yochem et al. 1998) to create the extrachromosomal array used for mosaic analysis. Animals that had lost the array completely were Peg due to nrf-6(sa367) and Muv (multivulva) due to lin-15(n765ts). Mosaic animals that lost the extrachromosomal array in the E lineage (intestine) were picked by their Peg and non-Muv phenotypes. Mosaic animals that lost the array in the AB lineage (hypodermis, including nose hypodermis, neurons, etc.) were picked by their non-Peg and partial Muv phenotypes. Candidate mosaic animals were scored for GFP expression in the nose hypodermis and intestine using a dissecting microscope with a UV lamp attachment and were then scored for the Nrf phenotype after incubation in 1.0 mg/ml fluoxetine for 10 min. The GFP expression pattern was then confirmed using a compound microscope with Nomarski optics and epifluorescence.
Molecular biology:
nrf-5(sa513); lin-15(n756ts) animals were rescued for both the Nrf and Peg phenotypes by cosmid pools containing F55B12, by F55B12 alone, or by pRC28, a 4.5-kb subclone containing the predicted gene F55B12.5 and 1.8 kb of promoter. Bulk PCR products of F55B12.5 from wild-type and nrf-5(sa513) genomic DNA were sequenced using an ABI automated sequencer and the sa513 mutation was confirmed by sequencing on both strands.
nrf-5 cDNAs yk56b12 and yk32e7 were obtained from the DNA Data Bank of Japan and were sequenced to confirm intron-exon boundaries of the F55B12.5 Genefinder predicted gene. From these sequences, we determined that an earlier prediction was missing a 75-bp intron in predicted exon 8 that was spliced out in both yk32e7 and yk56b12. In addition, we determined that exon 4 is spliced as predicted in both cDNAs. The yk32e7 cDNA is almost full length; it terminates 7 bp downstream of the predicted ATG of F55B12.5. Subsequent to our sequencing efforts, additional overlapping ESTs (yk1134g07, yk1436a01, yk1440f07, etc.) sequenced by the Genome Consortium and documented on WormBase (release WS138) confirmed that the F55B12.5-predicted ATG codon corresponds to the true translation initiation site of nrf-5. Both yk32e7 and yk56b12 have 89 nt 3′-UTRs followed by a poly(A) tail.
The nrf-5∷gfp plasmid pRC34 was created by inserting the GFP coding region from pPD95.67 (A. Fire) in a SnaBI site (generated by a silent mutation) at Val548 in the nrf-5-rescuing plasmid pRC28 (see above). nrf-5(sa513); lin-15(n756ts) animals were injected with pRC34 (25 ng/μl), pbLH98 (60 ng/μl), and pBluescript (Stratagene, La Jolla, CA) (78 ng/μl, carrier), and transgenic lines were analyzed for rescue of nrf-5 and GFP expression. An intestinal-specific version of the nrf-6∷gfp-rescuing plasmid pRC17 (Choy and Thomas 1999) was made by deleting a 1-kb NotI-EcoRI fragment of the promoter region followed by Klenow enzyme filling and religation.
RESULTS
Nrf–Nrf double mutants:
We previously found that Nrf mutants are only partially fluoxetine resistant (Choy and Thomas 1999). One explanation for this observation is that they play similar roles in conferring sensitivity to fluoxetine-induced nose contraction, but act in parallel. If this were the case, then Nrf–Nrf double mutants should have increased resistance to fluoxetine-induced nose contraction when compared to the single mutants. Alternatively, the Nrf genes could act in a common signaling pathway. In this case, Nrf–Nrf double mutants should have similar resistance as single null mutants. To distinguish between these hypotheses, we constructed various Nrf–Nrf double mutants and assayed their response to fluoxetine.
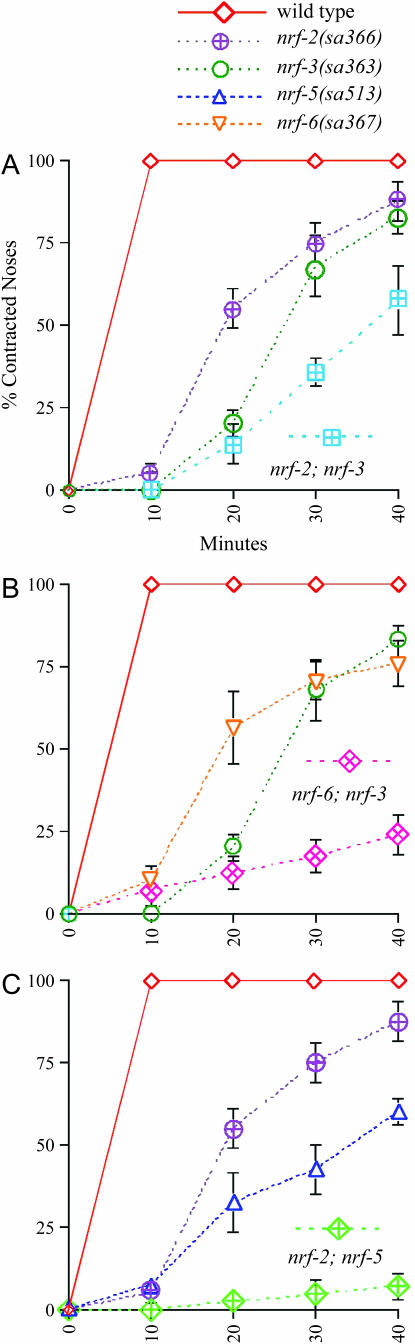
We previously found that all pairwise combinations of double mutants between null alleles of the three Nrf Peg genes, nrf-5, nrf-6, and ndg-4, were indistinguishable from the respective single mutants for both the Nrf and the Peg phenotype (Choy and Thomas 1999). These results suggest that nrf-5, nrf-6, and ndg-4 function in the same pathway to confer sensitivity to fluoxetine-induced nose contraction and that elimination of the function of any of these three genes is sufficient to eliminate the function of the entire pathway. To test whether the Nrf non-Peg genes nrf-2 and nrf-3 act in a common pathway, we constructed nrf-2; nrf-3 double mutants and assayed their Nrf phenotype. We found that the nrf-2; nrf-3 double mutant was slightly more resistant to fluoxetine-induced nose contraction than either of the single mutants (Figure 1A). Finally, to determine whether the Nrf Peg genes and the Nrf non-Peg genes act to confer fluoxetine sensitivity by different pathways, we constructed double mutants between these two classes. We constructed all pairwise combinations among Nrf Pegs nrf-5, nrf-6, and ndg-4 with the Nrf non-Pegs nrf-2 and nrf-3 (with the exception of nrf-2 nrf-6, because these genes are closely linked to each other on chromosome II). We found that all these double mutants were much more resistant to fluoxetine than the respective single mutants (Figure 1, B and C; data not shown). This suggests that the Nrf Peg genes and the Nrf non-Peg genes act in different pathways to confer fluoxetine sensitivity. The common secondary phenotypes among these two classes of Nrf mutants (Peg or non-Peg) further supports this interpretation.
Figure 1.
Nrf–Nrf double mutants are enhanced for their Nrf phenotype. Time courses of nose contraction for various double mutants and their respective single-mutant controls in 1.0 mg/ml fluoxetine. (A) Double mutant between Nrf non-Peg mutants nrf-2 and nrf-3. The double mutant is slightly more resistant than the respective single mutants. (B) Double mutant between Nrf Peg mutant nrf-6 and Nrf non-Peg mutant nrf-3. The double mutant is clearly more resistant than either of the single mutants. (C) Double mutant between Nrf non-Peg mutant nrf-2 and Nrf Peg mutant nrf-5. The double mutant is again clearly more resistant than the single mutants. Similar results were found with other Nrf Peg–Nrf non-Peg double-mutant combinations as described in the text. Each plot represents the average of at least four trials of 10 animals each. Some error bars are smaller than the plot symbols.
nrf-6 site of action:
nrf-6 and ndg-4 mutant animals have multiple phenotypes (Nrf and Peg) and nrf-6∷gfp and ndg-4∷gfp fusions are expressed in hypodermis and intestine (Choy and Thomas 1999). Because the mutants were isolated for a phenotype that affects the nose muscles and the nrf-6 and ndg-4 hypodermal expression is highest in the hypodermal cells surrounding those muscles (hyp 4 and hyp 5), we hypothesized that expression of these genes in the nose hypodermal cells was required for fluoxetine-induced nose contraction. In contrast, the pale egg phenotype of nrf-6 and ndg-4 results from a defect in yolk protein transport. In C. elegans, yolk proteins are normally synthesized in the intestine, secreted into the pseudocoelomic space, and then taken up into oocytes prior to fertilization (Kimble and Sharrock 1983; Grant and Hirsh 1999). nrf-6 and ndg-4 mutants accumulate yolk globules in the pseudocoelomic fluid, suggesting that yolk transport into oocytes, rather than yolk synthesis, is defective. However, this observation is also consistent with a defect in some aspect of yolk synthesis or transport from the intestine that decreases efficiency of yolk protein uptake into oocytes. It therefore seemed reasonable that nrf-6 and ndg-4 expression in the intestine is required for efficient yolk transport into oocytes.
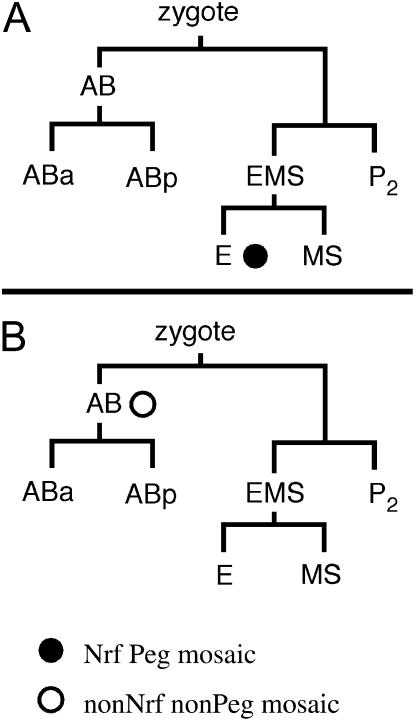
We used mosaic analysis to determine the site of action of nrf-6 for each of the two nrf-6 mutant phenotypes. C. elegans extrachromosomal arrays are lost at a low frequency during mitosis and can therefore be used to generate mosaic animals (Stinchcomb et al. 1985). We expressed an extrachromosomal array composed of sur-5∷gfp and a genomic cosmid containing nrf-6(+) in a nrf-6(sa367) mutant strain. sur-5∷gfp is expressed in nearly every cell in C. elegans and can therefore be used as a cell-autonomous marker (Yochem et al. 1998). The intestinal and hypodermal cell types are conveniently separated in the C. elegans cell lineage: the nose hypodermal cells, including hyp 4 and hyp 5, are derived from the AB blastomere, whereas the intestinal cells are derived from the E blastomere (Figure 2; Sulston et al. 1983).
Figure 2.
nrf-6 mosaic analysis. Circles indicate the cell division in which the nrf-6(+) extrachromosomal array was lost in the mosaic animals. The solid circle indicates Nrf Peg mosaics; the open circle indicates non-Nrf non-Peg mosaics. (A) Lineage diagram of Nrf Peg mosaics. All intestinal cells (and only intestinal cells) are derived from the E blastomere. Ten mosaics of this class were isolated. (B) Lineage diagram of non-Nrf non-Peg mosaic animals. All nose hypodermal cells including hyp 4 and hyp 5, as well as many other hypodermal and neuronal cells, are derived from the AB blastomere. Ten mosaics of this class were isolated: all were non-Peg and 9 were non-Nrf.
We identified 10 mosaic animals that had lost the extrachromosomal array in all intestinal cells but were still expressing GFP in most or all of the nose hypodermal cells hyp 4 and hyp 5 (see materials and methods). All 10 of these mosaics were both Nrf and Peg. We also isolated the reciprocal class of mosaic animals that had lost the extrachromosomal array in nose hypodermal cells but not in the intestine. Of 10 of these mosaics, 9 were non-Nrf and non-Peg (Figure 2). Taken together, the data from these two classes of mosaic animals indicate that, contrary to our expectations, nrf-6 expression is required in the intestine for fluoxetine-induced nose contraction as well as for efficient yolk transport. The significance of nrf-6 expression in the hypodermis remains unclear. We also performed additional experiments expressing nrf-6(+) under either an intestinal-specific truncated nrf-6 promoter or a hypodermal-specific dpy-7 promoter (Gilleard et al. 1997) in nrf-6(sa367) mutants. We found that intestinal-specific expression of nrf-6(+) rescued both the Nrf and the Peg defect of nrf-6 mutants, whereas hypodermal-specific expression did not rescue either of the defects (data not shown). The results of these tissue-specific expression experiments support the conclusions of our mosaic analysis. Although we did not perform mosaic analysis of ndg-4, nrf-6 and ndg-4 have identical mutant phenotypes and act in the same pathway by genetic criteria. Moreover, nrf-6 and ndg-4 encode homologous proteins and have the same expression pattern based on GFP fusions (Choy and Thomas 1999). We conclude that ndg-4 very likely also acts in the intestine to confer sensitivity to fluoxetine-induced nose contraction and to mediate efficient yolk uptake.
nrf-5 is a member of a family of secreted lipid-binding proteins:
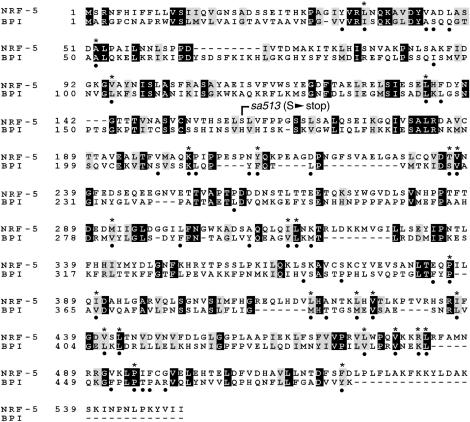
nrf-5 mutants have the same Nrf and Peg phenotypes as nrf-6 and ndg-4. We had previously shown, on the basis of genetic analysis, that nrf-5, nrf-6, and ndg-4 all act in the same pathway (Choy and Thomas 1999). Because nrf-6 and ndg-4 encoded novel proteins of unknown function, we sought to elucidate the function of the Nrf Peg genes by cloning nrf-5. We found that a cosmid subclone containing a single predicted gene (F55B12.5) rescued both the Nrf and the Peg phenotype of nrf-5 mutants (see materials and methods). We sequenced F55B12.5 genomic DNA from nrf-5(sa513) mutants and found a stop mutation at Ser159 (Figure 3). This early stop mutation is predicted to be a null allele. On the basis of these results, we conclude that nrf-5 is F55B12.5. We obtained two nrf-5 EST cDNAs from the C. elegans Genome Project and sequenced them to determine the intron-exon boundaries of F55B12.5 (see materials and methods). Using BLAST searches, we found that NRF-5 is homologous to four mammalian-secreted lipid-binding proteins: bactericidal permeability-increasing (BPI) protein (Gray et al. 1989; Beamer et al. 1997; Beamer 2003), lipopolysaccharide-binding protein (LBP; Fenton and Golenbock 1998; Iovine et al. 2002; Mulero et al. 2002), cholesteryl ester transfer protein (CETP; Bruce et al. 1998a; Kawano et al. 2000), and phospholipid transfer protein (PLTP; Huuskonen et al. 1999; Vakkilainen et al. 2002). These proteins have diverse functions, but share the ability to bind various lipids. NRF-5 is also similar to a number of predicted C. elegans proteins of unknown function (with corresponding orthologs in the related nematode C. briggsae) as well as several to predicted fish proteins (Figure 4). Interestingly, no convincing reciprocal match to nrf-5 was found in the Drosophila genome.
Figure 3.
Alignment of nrf-5 and human BPI protein. Solid boxes denote identical amino acids; shaded boxes denote similarities. The position of the mutation S159 to stop in sa513 is indicated. Solid dots below the alignment indicate 47 phospholipid contact residues based on BPI crystal structure (Beamer et al. 1997). Asterisks above the alignment indicate the 25 identical or similar residues in nrf-5 among these 47 contact residues.
Figure 4.
nrf-5 is homologous to a family of mammalian lipid-binding proteins. Dendrogram of nrf-5 and related proteins. NRF-5 and its three closest relatives in C. elegans and their reciprocal-best-match orthologs in C. briggsae are compared with a variety of relatives in vertebrates. In each name, the first small letters indicate the organism (ce, C. elegans; cb, C. briggsae; hs, Homo sapiens; mus, Mus musculus; trout, Oncorhynchus mykiss; carp, Cyprinus carpio; cod, Cadus morhua), the next segment is a commonly used literature name if it exists, and the final segment is a unique database identifier (in parentheses when it follows a common name).
Expression pattern of nrf-5:
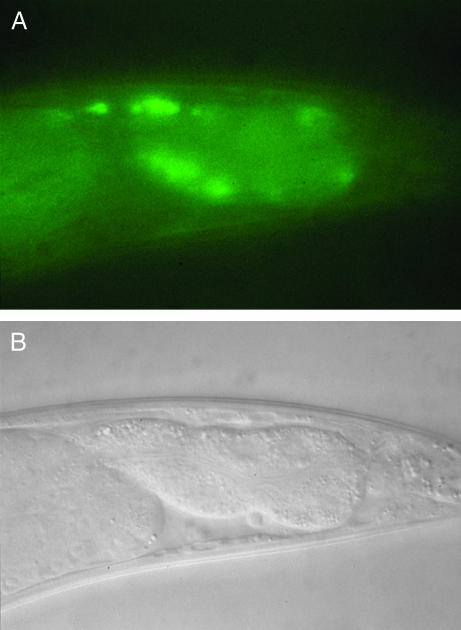
To determine the expression pattern of nrf-5, we inserted GFP (Chalfie et al. 1994) at the C-terminal end of nrf-5 in the genomic subclone used to rescue nrf-5 mutants (see materials and methods). We observed GFP fluorescence throughout the entire intestine of transgenic animals expressing this nrf-5∷gfp fusion (Figure 5; data not shown). No GFP expression was observed in any other tissue. Furthermore, the nrf-5∷gfp fusion rescued the Nrf and Peg phenotypes of nrf-5 mutants (data not shown). We found that nrf-5∷gfp fluorescence was concentrated at the basal membrane of the intestine (Figure 5). On the basis of its similarity to secreted lipid-binding proteins and the fact that it contains an N-terminal signal sequence (Nielsen et al. 1997), we conclude that NRF-5 is secreted. The concentration of nrf-5∷gfp fluorescence at the basal membrane of the intestine suggests that NRF-5 protein is secreted from the intestine into the pseudocoelomic fluid. We did not observe GFP fluorescence in the pseudocoelomic fluid itself; however, one possibility is that the level of secreted nrf-5∷gfp is too low to detect in this manner, particularly against the background of fluorescence from the intestine.
Figure 5.
Expression pattern of nrf-5∷gfp fusion. (A) Fluorescence photograph of posterior intestine of adult hermaphrodite expressing nrf-5∷gfp fusion. Anterior is to the left. Fluorescence is concentrated at the basal membrane of intestinal cells, consistent with the homology of NRF-5 to secreted proteins. (B) Nomarski photograph corresponding to A.
DISCUSSION
Nrf Peg and Nrf non-Peg genes act in distinct pathways:
We found that double mutants between Nrf Peg genes (nrf-5, nrf-6, and ndg-4) and Nrf non-Peg genes (nrf-2 and nrf-3) had dramatically increased fluoxetine resistance when compared to respective single mutants. This result suggests that the Nrf Peg genes and the Nrf non-Peg genes act in distinct pathways to confer fluoxetine sensitivity in C. elegans. We also found that nrf-2; nrf-3 double mutants had slightly increased fluoxetine resistance when compared to nrf-2 and nrf-3 single mutants. The simple interpretation of this result is that nrf-2 and nrf-3 also function in different pathways to confer fluoxetine sensitivity. However, an alternative possibility is that nrf-2 and nrf-3 are both partial loss-of-function mutations and that the small enhancement in the nrf-2; nrf-3 double mutant is explained by an additive effect of two mutations in the same pathway. Without information about the molecular nature of the single alleles of nrf-2 and nrf-3, we cannot distinguish between these two interpretations.
nrf-5 may encode a secreted lipid-binding protein:
We found that nrf-5 is homologous to a family of secreted lipid-binding proteins including BPI, LBP, CETP, and PLTP. Members of this family have been identified in a wide range of organisms, including humans, chickens, and fish (BLAST; Beamer et al. 1998). BPI and LBP both bind to lipopolysaccharide, a component of bacterial cell walls, and function in the immune system during bacterial infection (Beamer et al. 1999; Iovine et al. 2002; Weiss 2003). CETP and PLTP are involved in metabolism and transport of cholesterol and other lipids in the bloodstream (Bruce et al. 1998b; Kawano et al. 2000; Desrumaux et al. 2001). The only common function among members of this diverse family is the ability to bind to large hydrophobic molecules such as lipids and cholesterol (Bruce et al. 1998a; Guyard-Dangremont et al. 1999).
The structure of BPI at atomic resolution has been determined by X-ray crystallography (Beamer et al. 1997). One molecule of the phospholipid phosphatidyl choline was bound to BPI in each of two hydrophobic-binding pockets. It was proposed that other phospholipids would be able to bind in this pocket as well, consistent with the relatively low specificity of lipid substrate binding among members of the family (Beamer et al. 1997). From the BPI crystal structure, 47 of 456 amino acids were predicted to interact with the bound phospholipids. nrf-5 is 35% similar to BPI overall, but it is 53% similar among these 47 predicted lipid contact residues (Figure 3). This degree of conservation is similar to other family members such as CETP and PLTP (36% and 42% similar to BPI overall and 49% and 57% similar among lipid contact residues, respectively). Therefore, it is likely that nrf-5 binds large hydrophobic molecules in a similar binding pocket.
Models for nrf-5, nrf-6, and ndg-4 function:
The Nrf Peg mutants nrf-5, nrf-6, and ndg-4 represent a specific subclass of the mutants isolated in a screen for Nrf mutants (Choy and Thomas 1999). The nrf-6 and ndg-4 gene products, as with all members of this protein family, contain a consensus acyl transferase domain identified by clusters of orthologous groups analysis (National Center for Biotechnology Information). Interestingly, one Drosophila member of the NRF-6/NDG-4 family has been reported to be expressed in embryonic and adult nervous tissue (Dzitoyeva et al. 2003). On the basis of mutant phenotypes and molecular identities of the C. elegans genes, several models are possible to explain how nrf-5, nrf-6, and ndg-4 function in wild-type animals to confer sensitivity to fluoxetine-induced nose contraction and to mediate uptake of yolk proteins. We outline the details of two such models in the following sections.
Drug and yolk transport model:
In this model, NRF-6 and NDG-4 functions in the intestine are required for the function of NRF-5, a protein that is secreted into the pseudocoelomic fluid. This is consistent with the common phenotypes of mutants in these three genes, as well as with our finding that NRF-6 functions in the intestine. In this model, NRF-5 binds and transports two different substances, each of which explains one of the mutant phenotypes (Nrf and Peg). First, NRF-5 binds to yolk proteins and is required to transport or present them to oocytes for endocytosis. C. elegans yolk proteins are lipoproteins that are composed of ∼15% lipid (Sharrock et al. 1990). NRF-6 and NDG-4 may participate in transport or fatty acylation of yolk proteins at the site of the basal intestinal membrane. Since NRF-5 is homologous to lipid-binding proteins, one possibility is that it binds to secreted yolk lipoproteins via their lipid moieties. This could explain the yolk accumulation that is seen in Nrf Peg mutants. However, nrf-5∷gfp and nrf-6∷gfp were expressed in males, which do not produce yolk, and both nrf-5 and nrf-6 males are Nrf, indicating that the Nrf Peg genes have additional functions other than mediating yolk transport in hermaphrodites. The second function of NRF-5 in this model is to bind to fluoxetine and transport it to target tissues where the drug induces nose contraction. This function is consistent with the homology of NRF-5 to the mammalian lipid transfer proteins CETP and PLTP (see above). Both the selective serotonin reuptake inhibitors and tricyclic antidepressants to which Nrf mutants are resistant are hydrophobic. NRF-5 may bind both types of molecules in its putative hydrophobic binding pocket, consistent with the low binding specificity observed for mammalian members of this family (Bruce et al. 1998a). This model does not strictly require fluoxetine to bind to NRF-5 to induce nose contraction, since presumed nrf-5 null mutants are only partially resistant. One possibility is that NRF-5 binding and transport of fluoxetine expedites a process that happens at a slower rate in Nrf Peg null mutants. Alternatively, other proteins, perhaps other members of the putative NRF-5 lipid-binding protein family, may be partially redundant with NRF-5.
Lipid metabolism defect model:
In this second model, NRF-5, NRF-6, and NDG-4 mediate some aspect of lipid metabolism and a defect in this metabolic process is responsible for both the Nrf and the Peg phenotype of these mutants. For example, NRF-5, NRF-6, and NDG-4 could be involved in the transport or modification of particular lipids from the intestine, affecting their availability to other tissues through the pseudocoelomic fluid. An example is given by the human acyl transferase ACAT, which influences the availability of cholesterol by reversible esterification to fatty acids (reviewed in Chang et al. 1997). This enzyme is unrelated to NRF-6 and NDG-4 (not shown). The yolk uptake defect of these mutants could be explained by the requirement of a particular lipid component in yolk proteins for their efficient uptake into oocytes. In the mutants, this lipid component is absent or aberrant, thereby leading to accumulation of yolk in the pseudocoelomic space. This is consistent with work demonstrating that yolk protein uptake by oocytes requires the lipoprotein receptor encoded by rme-2 (Grant and Hirsh 1999).
A defect in lipid metabolism could also explain the Nrf mutant phenotype of nrf-5, nrf-6, and ndg-4. Changes in the lipid composition of a muscle or neuronal cell membrane could directly or indirectly alter its excitable properties. For example, several neurotransmitter receptors, including nicotinic acetylcholine receptors and glutamate receptors, have been shown to be modulated by the lipid composition of the membranes in which they are imbedded (Sunshine and McNamee 1994; Gagne et al. 1996). Alternatively, changes in lipid composition could result in changes in lipid secondary messenger pools (Graber et al. 1994; Leevers et al. 1999) in a signaling pathway that is responsible for fluoxetine-induced nose contraction. It is important to note that Nrf mutants do not have a nonspecific decrease in nose muscle excitability because they are fully sensitive to several nonantidepressant drugs that induce muscle contraction (Choy and Thomas 1999).
Although the suggestion that changes in lipid metabolism could result in neuromuscular defects is unusual, there are at least two precedents for similar mutant phenotypes. First, C. elegans fat-3 fatty acid desaturase mutants are defective in the biosynthesis of long chain polyunsaturated fatty acids and have uncoordinated movement defects (Lesa et al. 2003; Watts et al. 2003). Second, Drosophila easily shocked mutants were isolated on the basis of a bang-sensitive paralytic phenotype, and easily shocked encodes an ethanolamine kinase, an enzyme involved in the biosynthesis of the ubiquitous phosphatidyl ethanolamine (Pavlidis et al. 1994). The exact mechanisms by which mutations in these two genes confer these mutant phenotypes have not yet been fully elucidated, but these examples clearly demonstrate that defects in lipid metabolism can result in unexpected neuromuscular defects and support the idea that the Nrf phenotype of nrf-5, nrf-6, and ndg-4 mutants could be explained by similar defects.
Acknowledgments
We thank Margaret Niedenthal at Eli Lilly for fluoxetine; Alan Coulson for cosmids; the Genome Sequencing Centers for sequence information; Andrew Fire for GFP vectors; Jennifer Watts for data prior to publication; Iain Johnstone for the dpy-7 promoter construct; and Yuji Kohara, Susan Haynes, and Steve Williams for cDNAs. Some of the strains used in this study were obtained from the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) National Center for Research Resources. R.K.M.C. was supported by a Predoctoral Fellowship from the Howard Hughes Medical Institute. This work was supported by NIH grant R01-MH58916 to J.H.T.
References
- Beamer, L. J., 2003. Structure of human BPI (bactericidal/permeability-increasing protein) and implications for related proteins. Biochem. Soc. Trans. 31: 791–794. [DOI] [PubMed] [Google Scholar]
- Beamer, L. J., S. F. Carroll and D. Eisenberg, 1997. Crystal structure of human BPI and two bound phospholipids at 2.4 angstrom resolution. Science 276: 1861–1864. [DOI] [PubMed] [Google Scholar]
- Beamer, L. J., D. Fischer and D. Eisenberg, 1998. Detecting distant relatives of mammalian LPS-binding and lipid transport proteins. Protein Sci. 7: 1643–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beamer, L. J., S. F. Carroll and D. Eisenberg, 1999. The three-dimensional structure of human bactericidal/permeability-increasing protein: implications for understanding protein-lipopolysaccharide interactions. Biochem. Pharmacol. 57: 225–229. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, C., L. J. Beamer and A. R. Tall, 1998. a The implications of the structure of the bactericidal/permeability-increasing protein on the lipid-transfer function of the cholesteryl ester transfer protein. Curr. Opin. Struct. Biol. 8: 426–434. [DOI] [PubMed] [Google Scholar]
- Bruce, C., R. A. Chouinard, Jr. and A. R Tall, 1998. b Plasma lipid transfer proteins, high-density lipoproteins, and reverse cholesterol transport. Annu. Rev. Nutr. 18: 297–330. [DOI] [PubMed] [Google Scholar]
- Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward and D. C. Prasher, 1994. Green fluorescent protein as a marker for gene expression. Science 263: 802–805. [DOI] [PubMed] [Google Scholar]
- Chang, T. Y., C. C. Y. Chang and D. Cheng, 1997. Acyl-coenzyme A: cholesterol acyltransferase. Annu. Rev. Biochem. 66: 613–638. [DOI] [PubMed] [Google Scholar]
- Choy, R. K. M., and J. H. Thomas, 1999. Fluoxetine-resistant mutants in C. elegans define a novel family of transmembrane proteins. Mol. Cell 4: 143–152. [DOI] [PubMed] [Google Scholar]
- Clark, S. G., X. Lu and H. R. Horvitz, 1994. The Caenorhabditis elegans locus lin-15, a negative regulator of a tyrosine kinase signaling pathway, encodes two different proteins. Genetics 137: 987–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent, J. A., M. M. Smith, D. K. Vassilatis and L. Avery, 2000. The genetics of ivermectin resistance in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 97: 2674–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrumaux, C., C. Labeur, A. Verhee, J. Tavernier, J. Vandekerckhove et al., 2001. A hydrophobic cluster at the surface of the human plasma phospholipid transfer protein is critical for activity on high density lipoproteins. J. Biol. Chem. 276: 5908–5915. [DOI] [PubMed] [Google Scholar]
- Dzitoyeva, S., N. Dimitrijevic and H. Manev, 2003. Identification of a novel Drosophila gene, beltless, using injectable embryonic and adult RNA interference (RNAi). BMC Genomics 4: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton, M. J., and D. T. Golenbock, 1998. LPS-binding proteins and receptors. J. Leukoc. Biol. 64: 25–32. [DOI] [PubMed] [Google Scholar]
- Fleming, J. T., M. D. Squire, T. M. Barnes, C. Tornoe, K. Matsuda et al., 1997. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J. Neurosci. 17: 5843–5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne, J., C. Giguere, G. Tocco, M. Ohayon, R. F. Thompson et al., 1996. Effect of phosphatidylserine on the binding properties of glutamate receptors in brain sections from adult and neonatal rats. Brain Res. 740: 337–345. [DOI] [PubMed] [Google Scholar]
- Gilleard, J. S., J. D. Barry and I. L. Johnstone, 1997. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Mol. Cell. Biol. 17: 2301–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber, R., C. Sumida and E. A. Nunez, 1994. Fatty acids and cell signal transduction. J. Lipid Mediat. Cell Signal. 9: 91–116. [PubMed] [Google Scholar]
- Grant, B., and D. Hirsh, 1999. Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol. Biol. Cell 10: 4311–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, P. W., G. Flaggs, S. R. Leong, R. J. Gumina, J. Weiss et al., 1989. Cloning of the cDNA of a human neutrophil bactericidal protein. Structural and functional correlations. J. Biol. Chem. 264: 9505–9509. [PubMed] [Google Scholar]
- Guyard-Dangremont, V., V. Tenekjian, V. Chauhan, S. Walter, P. Roy et al., 1999. Immunochemical evidence that cholesteryl ester transfer protein and bactericidal/permeability-increasing protein share a similar tertiary structure. Protein Sci. 8: 2392–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huuskonen, J., G. Wohlfahrt, M. Jauhiainen, C. Ehnholm, O. Teleman et al., 1999. Structure and phospholipid transfer activity of human PLTP: analysis by molecular modeling and site-directed mutagenesis. J. Lipid Res. 40: 1123–1130. [PubMed] [Google Scholar]
- Iovine, N., J. Eastvold, P. Elsbach, J. P. Weiss and T. L. Gioannini, 2002. The carboxyl-terminal domain of closely related endotoxin-binding proteins determines the target of protein-lipopolysaccharide complexes. J. Biol. Chem. 277: 7970–7978. [DOI] [PubMed] [Google Scholar]
- Kawano, K., S.-C. Qin, M. Lin, A. R. Tall and X.-C. Jiang, 2000. Cholesteryl ester transfer protein and phospholipid transfer protein have nonoverlapping functions in vivo. J. Biol. Chem. 275: 29477–29481. [DOI] [PubMed] [Google Scholar]
- Kimble, J., and W. J. Sharrock, 1983. Tissue-specific synthesis of yolk proteins in Caenorhabditis elegans. Dev. Biol. 96: 189–196. [DOI] [PubMed] [Google Scholar]
- Leevers, S. J., B. Vanhaesebroeck and M. D. Waterfield, 1999. Signaling through phosphoinositide 3-kinases: the lipids take centre stage. Curr. Opin. Cell Biol. 11: 219–225. [DOI] [PubMed] [Google Scholar]
- Lesa, G. M., M. Palfreyman, D. H. Hall, M. T. Clandinin, C. Rudolph et al., 2003. Long chain polyunsaturated fatty acids are required for efficient neurotransmission in C. elegans. J. Cell Sci. 116: 4965–4975. [DOI] [PubMed] [Google Scholar]
- Lewis, J. A., C. H. Wu, H. Berg and J. H. Levine, 1980. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics 95: 905–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. G., A. Alfonso, M. Nguyen, J. A. Crowell, C. D. Johnson et al., 1996. A genetic selection for Caenorhabditis elegans synaptic transmission mutants. Proc. Natl. Acad. Sci. USA 93: 12593–12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, K. G., M. D. Emerson, J. R. McManus and J. B. Rand, 2000. RIC-8 (Synembryn): a novel conserved protein that is required for Gqα signaling in the C. elegans nervous system. Neuron 27: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero, J. J., B. J. Boyle, S. Bradley, J. M. Bright, S. T. Nelken et al., 2002. Three new human members of the lipid transfer/lipopolysaccharide binding protein family (LT/LBP). Immunogenetics 54: 293–300. [DOI] [PubMed] [Google Scholar]
- Nguyen, M., A. Alfonso, C. D. Johnson and J. B. Rand, 1995. Caenorhabditis elegans mutants resistant to inhibitors of acetylcholinesterase. Genetics 140: 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, H., J. Engelbrecht, S. Brunak and G. von Heijne, 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1–6. [DOI] [PubMed] [Google Scholar]
- Pavlidis, P., M. Ramaswami and M. A. Tanouye, 1994. The Drosophila easily shocked gene: a mutation in a phospholipid synthetic pathway causes seizure, neuronal failure, and paralysis. Cell 79: 23–33. [DOI] [PubMed] [Google Scholar]
- Schafer, W. R., 1999. How do antidepressants work? Prospects for genetic analysis of drug mechanisms. Cell 98: 551–554. [DOI] [PubMed] [Google Scholar]
- Sharrock, W. J., M. E. Sutherlin, K. Leske, T. K. Cheng and T. Y. Kim, 1990. Two distinct yolk lipoprotein complexes from Caenorhabditis elegans. J. Biol. Chem. 265: 14422–14431. [PubMed] [Google Scholar]
- Stinchcomb, D. T., J. E. Shaw, S. H. Carr and D. Hirsh, 1985. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 5: 3484–3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J.E., E. Schierenberg, J. G. White and J. N. Thomson, 1983. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100: 64–119. [DOI] [PubMed] [Google Scholar]
- Sunshine, C., and M. G. McNamee, 1994. Lipid modulation of nicotinic acetylcholine receptor function: the role of membrane lipid composition and fluidity. Biochim. Biophys. Acta 1191: 59–64. [DOI] [PubMed] [Google Scholar]
- Vakkilainen, J., M. Jauhiainen, K. Ylitalo, I. O. Nuotio, J. S. A. Viikari et al., 2002. LDL particle size in familial combined hyperlipidemia: effects of serum lipids, lipoprotein-modifying enzymes, and lipid transfer proteins. J. Lipid Res. 43: 598–603. [PubMed] [Google Scholar]
- Watts, J. L., E. Phillips, K. R. Griffing and J. Browse, 2003. Deficiencies in C20 polyunsaturated fatty acids cause behavioral and developmental defects in Caenorhabditis elegans fat-3 mutants. Genetics 163: 581–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, J., 2003. Bactericidal/permeability-increasing protein (BPI) and lipopolysaccharide-binding protein (LBP): structure, function and regulation in host defence against Gram-negative bacteria. Biochem. Soc. Trans. 31: 785–790. [DOI] [PubMed] [Google Scholar]
- Yochem, J., T. Gu and M. Han, 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]