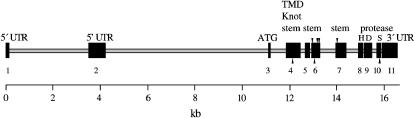
Figure 1.
Sb-sbd genomic structure. Sizes of exons and introns are represented to scale and indicated in kilobases at the bottom. Exons are shown as solid bars with exon number below. Features from the cDNA (5′- and 3′-UTR, ATG translation start) and amino acid sequence are indicated above the exons in which they are found. The protease domain is split among the last four exons. Each of the amino acids forming the catalytic triad (H, D, S) is encoded in a separate exon, indicated by marks above the appropriate exon. The transmembrane signal/anchor (TMD), disulfide knot (knot), and the first part of the stem are in exon 4. See text and Figure 2 for further description of protein domains. Four polymorphisms that result in a change in amino acid sequence are found in exons 6 and 7 and are indicated by marks above the line. In exon 6, an in-frame deletion of CAG in the Oregon-R strain reduces a string of seven glutamines to six between amino acids 287 and 293. An A-to-T change at nucleotide 1910 (cDNA numbering) results in a serine-for-threonine substitution at amino acid 349 within a serine/threonine (S/T)-rich region. At nucleotide 1924, a C-to-T change replaces a proline with a serine at amino acid 354. In exon 7, a third wild-type strain (the progenitor for the sbd201 mutant) deletes nine nucleotides inframe, resulting in the deletion of one unit of a tandem repeat of threonine, threonine, serine (TTS), between amino acids 410 and 416 in an S/T-rich region. Nucleotide differences between the Oregon-R and y; cn bw sp strains that produce no amino acid changes are found in exon 4 (nt 1150), exon 6 (nt1859), and exon 10 (nt 2198) and are indicated by marks below the exons.