Abstract
In many biological systems, especially bacteria and unicellular eukaryotes, rates of synonymous and nonsynonymous nucleotide divergence are negatively correlated with the level of gene expression, a phenomenon that has been attributed to natural selection. Surprisingly, this relationship has not been examined in many important groups, including the unicellular model organism Chlamydomonas reinhardtii. Prior to this study, comparative data on protein-coding sequences from C. reinhardtii and its close noninterfertile relative C. incerta were very limited. We compiled and analyzed protein-coding sequences for 67 nuclear genes from these taxa; the sequences were mostly obtained from the C. reinhardtii EST database and our C. incerta EST data. Compositional and synonymous codon usage biases varied among genes within each species but were highly correlated between the orthologous genes of the two species. Relative rates of synonymous and nonsynonymous substitution across genes varied widely and showed a strong negative correlation with the level of gene expression estimated by the codon adaptation index. Our comparative analysis of substitution rates in introns of lowly and highly expressed genes suggests that natural selection has a larger contribution than mutation to the observed correlation between evolutionary rates and gene expression level in Chlamydomonas.
IN diverse biological lineages the degree of nonrandom usage of synonymous codons and the rate of evolutionary change of a gene are often related to its level of expression. For example, in bacteria, yeast, Caenorhabditis elegans, Drosophila, and Arabidopsis, codon usage bias of genes appears to be positively correlated with the level of gene expression (Gouy and Gautier 1982; Sharp et al. 1986; Stenico et al. 1994; Chiapello et al. 1998; Duret and Mouchiroud 1999) and it is generally accepted that this is the result of elevated selection for a preferred set of codons that enhance translational efficiency in highly expressed genes (for reviews see Akashi and Eyre-Walker 1998; Akashi 2001). Moreover, in bacteria (Sharp and Li 1987a; Sharp 1991; Smith and Eyre-Walker 2001) and yeast (Pal et al. 2001; Hirsh et al. 2005), rates of synonymous substitutions are negatively correlated with the level of gene expression and it is generally thought that this is the result of purifying selection for efficient translation of highly expressed genes, although variation in mutation rate has been invoked to explain this correlation in some cases (Berg and Martelius 1995; Eyre-Walker and Bulmer 1995; but see Ochman 2003). In some multicellular lineages, however, including mammals and Arabidopsis, a correlation between synonymous rate and gene expression level could not be established (Duret and Mouchiroud 2000; Wright et al. 2002, 2004) and while there is evidence supporting such a correlation in Drosophila (e.g., Powell and Moriyama 1997), this correlation has been questioned (Dunn et al. 2001; but see Bierne and Eyre-Walker 2003 and Marais et al. 2004).
Rates of nonsynonymous substitution, which typically show greater variation among genes than synonymous rates, also appear correlated with level of gene expression in diverse lineages including various bacteria, yeast, land plants, Drosophila, and mammals (Duret and Mouchiroud 2000; Pal et al. 2001; Wright et al. 2002, 2004; Marais et al. 2004; Rispe et al. 2004; Rocha and Danchin 2004; Subramanian and Kumar 2004; Lemos et al. 2005). The basis for the connection between gene expression and protein evolution is the subject of ongoing debate. It has been suggested that nonsynonymous sites in highly expressed genes are under enhanced selective constraint to optimize the speed and accuracy of protein synthesis (Akashi 2001, 2003). Alternatively, genes expressed broadly across tissues, which are also highly expressed, may be under enhanced functional constraint because their products need to function in diverse biochemical/biophysical environments (Hastings 1996) or because nonsynononymous changes in these genes affect a greater number of tissues and therefore have a greater impact on fitness (Duret and Mouchiroud 2000).
The green algae represent a major biological group of eukaryotes for which there has been no large-scale examination of evolutionary rates in nuclear protein-coding genes. Chlamydomonas reinhardtii represents an obvious candidate for such a study, as this green algal species has an extensive and annotated expressed sequence tag (EST) database and a genome project near completion. The nuclear genome of C. reinhardtii has an overall GC content of nearly 65% (Grossman et al. 2003) and the nucleus-encoded genes of this taxon have a preponderance of codons that end in G or C, although the codon usage bias varies considerably among genes (LeDizet and Piperno 1995). Correspondence analysis of relative synonymous codon usage (RSCU) values has established that C. reinhardtii genes are positioned along the first axis according to the level of gene expression as approximated by EST abundance (Naya et al. 2001). The relationship between codon usage and level of gene expression in C. reinhardtii is consistent with the observation that highly expressed genes exhibit lower values of the effective number of codons (Nc). Furthermore, genes with the highest number of EST matches use a preferred set of codons, which are rich in C at fourfold degenerate sites relative to those with only one EST match (Naya et al. 2001).
C. incerta SAG 7.73 is the closest known noninterfertile relative of C. reinhardtii (Schlösser 1976; Coleman and Mai 1997; Ferris et al. 1997; Liss et al. 1997; Pröschold et al. 2001), with the possible exception of Chlamydomonas sp. (CCAP 11/132) (Pröschold et al. 2005). Studies on the level of sequence divergence between C. incerta and C. reinhardtii are limited to three spliceosomal introns (Liss et al. 1997), two intergenic spacers (Coleman and Mai 1997), and a few protein-coding genes (Ferris et al. 1997). Clearly, an expanded analysis of orthologous gene sequences from these taxa is needed to gain a better view of the extent and causes of the variation in evolutionary divergence among genes in Chlamydomonas.
In this study we investigate the relationship between the rates of nucleotide divergence and the level of gene expression in C. reinhardtii and C. incerta. We compiled a greatly expanded data set of 67 protein-coding sequences by utilizing our C. incerta EST data prepared for the Protist EST Program (PEP) (http://megasun.bch.umontreal.ca/pepdb/pep.html), the C. reinhardtii EST database (http://www.chlamy.org), and GenBank (http://www4.ncbi.nlm.nih.gov). The specific objectives were (i) to compare synonymous codon usage in C. incerta with that of C. reinhardtii; (ii) to examine the diversity of synonymous and nonsynonymous substitution rates among genes between C. reinhardtii and C. incerta; (iii) to access the relationship, if any, between the evolutionary rates, both synonymous and nonsynonymous, and the level of gene expression as estimated by the codon adaptation index (CAI), which is a measure of the degree to which the synonymous codon usage of a gene matches that of highly expressed genes (Sharp and Li 1987b); and (iv) to compare the rate of nucleotide substitution in introns of genes with a low CAI to those with a high CAI.
MATERIALS AND METHODS
C. incerta strain and growth conditions:
C. incerta was obtained from the Sammlung von Algenkulturen, Göttingen (SAG), Germany, where it is listed as SAG 7.73 and placed under the name C. reinhardtii on the basis of morphological criteria and susceptibility to autolysin from the C. reinhardtii group (SAG, personal communication). Strain CC-1870 previously maintained at the Chlamydomonas Genetic Center, Duke University, now replaced with strain CC-3871, and strain UTEX 2607 maintained at the University of Texas at Austin, were all derived from SAG 7.73 and should be equivalent (Ferris et al. 1997). Cells were grown under continuous “cool white” fluorescent lighting at 24° in (i) Tris–acetate–phosphate (TAP) medium (Harris 1989) with rotary shaking and 25 μmol photons·m−2·s−1 photosynthetically active radiation (PAR) and (ii) high-salt (HS) medium (Harris 1989) bubbled with 1% CO2 in air and 130 μmol photons·m−2·s−1 PAR. Cells were harvested by centrifugation at 2000 × g at 4° when the cultures reached the late exponential phase of growth (OD750 = 0.3 and 0.7 for the TAP- and HS-grown cultures, respectively). An equivalent mass of cells from each of the two culture conditions was lysed in TRIzol (Invitrogen, Carlsbad, CA) and the lysates were combined for RNA isolation. A pellet of HS-grown cells was resuspended in Tris–EDTA buffer (20 mm Tris, 100 mm EDTA, 100 mm NaCl, pH 8) for the isolation of DNA (Maniatis et al. 1982) used in PCR analysis.
Generation of the C. incerta (SAG 7.73) genomic sequences:
PCR amplification of several C. incerta genes was performed (i) to confirm that C. incerta strain SAG 7.73 is the same as the strain CC-1870 used by Ferris et al. (1997) and (ii) to obtain intron sequences to be used in substitution rate analyses. With regard to the first objective, the sequence of a 456-nucleotide segment of C. incerta (SAG 7.73) mid confirmed that this fragment is identical to the corresponding region of the C. incerta (CC-1870/CC-3871) mid deposited in GenBank (accession no. AF002710). The PCR reaction was performed using the following primers: 5′-TAG CCA GGT TCC GGT TCA A-3′ and 5′-CCA TCT GTC GAC GCC AAG T-3′. To study the intron substitution level, partial genomic sequences of cblP, GapA, and Tpx of SAG 7.73 were obtained using perfect match primer sequences based on the corresponding cDNA sequences obtained in this study. A 2124-nucleotide sequence of C. incerta sfa, not available as a cDNA, was amplified using a set of primer sequences based on the C. reinhardtii and C. eugametos orthologs: 5′-TGG AGC AGG AGA AGC AG-3′ and 5′-CGC CTT CGT GTA GTC GTT G-3′. Finally, the genomic sequence for pdk was obtained from GenBank for both C. reinhardtii and C. incerta (supplemental Table S1 at http://www.genetics.org/supplemental/).
C. incerta ESTs:
C. incerta (SAG 7.73) cDNA library construction and EST sequencing was part of the Protist EST program (http://megasun.bch.umontreal.ca/pepdb/pep.html). Non-normalized and normalized C. incerta cDNA libraries were constructed by DNA Technologies (Gaithersburg, MD). Inserts were unidirectionally cloned between the EcoRV and NotI sites of the pcDNA3.1 vector (Invitrogen). Sequencing was carried out at the National Research Council, Institute for Marine Biosciences, Halifax, Canada, and >95% of the ESTs were sequenced from the 5′-end; ∼85% of the sequenced ESTs were from the regular library. A total of 5124 quality- and vector-trimmed ESTs with an average length of 395 nucleotides were clustered into 1388 unique sequences (i.e., clusters and singletons) and 589 of these unique sequences gave BLASTX (Altschul et al. 1990) hits (expectation value of cutoff of 10−5) against the GenBank nonredundant database. The automatic annotation procedure using AutoFACT (Koski et al. 2005) was used to define the names of the C. incerta gene products.
Sequence retrieval and alignments:
The data set analyzed comprises 67 genes (Table 1 and supplemental Table S1 at http://www.genetics.org/supplemental/). Sixty-one C. incerta gene sequences were retrieved from the PEP database. The corresponding C. reinhardtii homologs were identified by running a BLASTN search (Altschul et al. 1990) of C. incerta unique sequences against the C. reinhardtii EST database from ChlamyDB (http://www.chlamy.org). In our C. incerta (SAG 7.73) cDNA library data, we found one EST sequence corresponding to yptC1. This sequence is different from the one reported by Ferris et al. (1997) as being the yptC1 of C. incerta (CC-1870/CC-3871). Our sequence, confirmed by sequencing the PCR product of the genomic yptC1, shows 22 synonymous substitution differences relative to the C. reinhardtii yptC1 sequence reported in GenBank (accession no. U13168), while the sequence reported by Ferris et al. (1997) differs from the C. reinhardtii GenBank entry at only one synonymous site. In all analyses we used the C. incerta yptC1 sequence obtained by us.
TABLE 1.
Estimates of dS and dN between C. incerta (Ci) and C. reinhardtii (Cr) and other evolutionary parameters
|
Ci/Cr
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene product description | Gene name | Lc | dS | dN | dN/dS | GC% | GC3% | Nc | CAI |
| Guanine nucleotide-binding β-subunit-like protein | cblP | 277 | 0.025 | 0.001 | 0.056 | 65.0/65.0 | 94.6/94.9 | 24.2/24.1 | 0.90/0.89 |
| 60S ribosomal protein L11 | rpl11 | 169a | 0.083 | 0.005 | 0.054 | 64.3/64.5 | 93.5/94.7 | 25.1/24.8 | 0.87/0.88 |
| Cytochrome b6f /Rieske Fe-S protein (p) | PetC | 178 | 0.099 | 0.020 | 0.198 | 66.2/66.7 | 83.7/84.8 | 27.3/27.5 | 0.79/0.78 |
| Photosystem I subunit F (p) | PsaF | 215 | 0.109 | 0.016 | 0.142 | 66.4/67.1 | 87.4/87.0 | 27.0/27.4 | 0.79/0.79 |
| 40S ribosomal protein S15 | rps15 | 129a | 0.110 | 0.006 | 0.057 | 62.5/62.5 | 89.9/89.9 | 26.3/26.1 | 0.89/0.88 |
| Cytochrome b6f/4.6-kDa subunit (p) | PetM | 98a | 0.133 | 0.004 | 0.033 | 71.4/69.4 | 87.8/81.6 | 26.9/29.5 | 0.79/0.72 |
| Ferredoxin (p) | PetF | 125a | 0.136 | 0 | — | 67.5/67.5 | 86.4/86.4 | 26.4/26.5 | 0.84/0.84 |
| ATP synthase subunit II (p) | AtpG | 208a | 0.136 | 0.010 | 0.071 | 65.7/65.4 | 90.9/88.9 | 26.4/27.0 | 0.80/0.80 |
| Translation initiation factor | Eif4A | 148 | 0.139 | 0 | — | 64.0/63.3 | 95.3/93.2 | 24.9/25.8 | 0.85/0.83 |
| Apoplastocyanin (p) | PetE | 144a | 0.139 | 0.015 | 0.106 | 66.0/66.4 | 83.3/84.0 | 26.6/25.7 | 0.80/0.82 |
| Glyceraldehyde-3-phosphate dehydrogenase (p) | GapA | 369 | 0.144 | 0.009 | 0.065 | 64.5/65.1 | 91.1/93.2 | 24.9/24.1 | 0.87/0.89 |
| NADH:ubiquinone oxidoreductase 49-kDa subunit (m) | Nad7 | 276 | 0.149 | 0.013 | 0.086 | 67.8/67.4 | 93.1/92.8 | 27.4/28.6 | 0.72/0.71 |
| Unknown lumenal polypeptide (p) | 116a | 0.155 | 0.016 | 0.105 | 69.3/67.6 | 89.3/86.4 | 24.2/25.2 | 0.82/0.83 | |
| 60S ribosomal protein L31 | rpl31 | 115a | 0.168 | 0.010 | 0.058 | 61.7/61.5 | 93.0/93.0 | 27.6/28.2 | 0.73/0.76 |
| Dynein light chain outer arm 8-kDa | fla14 | 90a | 0.170 | 0 | — | 58.1/57.5 | 90.0/87.8 | 41.7/46.8 | 0.73/0.70 |
| 16-kDa membrane protein (p) | 125a | 0.170 | 0.006 | 0.037 | 68.0/68.3 | 92.0/92.8 | 25.0/24.0 | 0.86/0.88 | |
| Apocytochrome c (m) | Cyc | 111a | 0.171 | 0.007 | 0.043 | 63.1/62.8 | 82.9/81.1 | 32.5/31.1 | 0.72/0.71 |
| 14-3-3-like protein | erb14 | 197 | 0.177 | 0.006 | 0.034 | 61.4/61.1 | 89.8/88.3 | 30.4/32.7 | 0.70/0.66 |
| ATP synthase F0 subunit 9 (m) | Atp9 | 158a | 0.178 | 0 | — | 68.1/67.1 | 86.8/83.6 | 25.6/28.4 | 0.75/0.73 |
| 40S ribosomal protein S3 | rps3a | 229a | 0.180 | 0.007 | 0.040 | 64.6/63.5 | 87.3/85.6 | 28.8/29.3 | 0.80/0.77 |
| Oxygen-evolving enhancer protein 1/OEE1 (p) | Psb1 | 291a | 0.181 | 0.006 | 0.031 | 65.1/64.9 | 90.0/89.3 | 25.4/26.6 | 0.86/0.85 |
| ATP synthase Δ-subunit (p) | AtpD | 218a | 0.190 | 0.021 | 0.112 | 62.5/62.2 | 87.6/86.7 | 28.3/28.6 | 0.77/0.76 |
| Cytochrome c oxidase subunit II (m) | Cox2a | 137 | 0.193 | 0.010 | 0.051 | 67.9/68.1 | 81.8/81.0 | 28.9/29.4 | 0.68/0.67 |
| Elongation factor α-like | efl | 462a | 0.194 | 0.008 | 0.043 | 62.2/62.1 | 90.5/89.8 | 25.0/25.0 | 0.87/0.87 |
| Light-harvesting complex I protein (p) | LhcA | 240a | 0.196 | 0.025 | 0.128 | 65.8/66.4 | 87.9/88.7 | 27.3/26.0 | 0.80/0.84 |
| Thioredoxin peroxidase (p) | Tpx | 236 | 0.206 | 0.010 | 0.049 | 65.4/64.8 | 92.0/87.7 | 25.7/27.8 | 0.87/0.81 |
| Light-harvesting chlorophyll-a/b binding protein (p) | LhcII-1.3 | 256a | 0.211 | 0.010 | 0.053 | 68.3/67.5 | 95.3/93.4 | 22.6/23.9 | 0.93/0.90 |
| 14-3-3-like protein related | 258a | 0.218 | 0.003 | 0.014 | 62.7/62.0 | 89.2/86.8 | 29.9/30.6 | 0.74/0.72 | |
| Cytochrome c oxidase subunit II (m) | Cox2b | 152a | 0.220 | 0.011 | 0.048 | 62.3/62.1 | 88.8/88.2 | 33.8/31.1 | 0.68/0.68 |
| 40S ribosomal protein S8 | rps8a | 207a | 0.221 | 0.019 | 0.085 | 63.6/63.5 | 93.2/93.2 | 30.0/29.6 | 0.77/0.79 |
| Nucleic acid binding protein putative | nab1 | 225 | 0.230 | 0.011 | 0.049 | 70.8/71.6 | 85.8/88.0 | 35.1/32.9 | 0.61/0.62 |
| Ubiquinol-cytochrome c oxidoreductase/Rieske Fe-S protein (m) | RisP | 211 | 0.241 | 0.004 | 0.016 | 65.6/65.7 | 87.2/88.2 | 31.1/30.7 | 0.66/0.67 |
| ATP synthase γ-subunit (p) | AtpC | 232 | 0.248 | 0.020 | 0.080 | 64.5/63.2 | 96.6/93.5 | 23.5/23.1 | 0.91/0.90 |
| G-strand binding protein 1/telomere binding | gbp1 | 219a | 0.251 | 0.008 | 0.031 | 69.3/69.4 | 84.0/83.1 | 32.2/32.5 | 0.71/0.71 |
| S-adenosyl-l-homocystein hydrolase | sahH | 256 | 0.274 | 0.014 | 0.050 | 65.9/65.5 | 93.0/91.2 | 24.9/24.7 | 0.88/0.87 |
| Photosystem I subunit N (p) | PsaN | 138a | 0.280 | 0.006 | 0.021 | 66.4/66.4 | 91.3/89.9 | 26.1/26.1 | 0.87/0.86 |
| GTP-binding YPTC1 protein | yptC1 | 202a | 0.289 | 0.000 | — | 58.0/58.8 | 84.1/86.6 | 37.3/36.3 | 0.63/0.65 |
| Oxygen-evolving enhancer protein 3/OEE3 (p) | Psb3 | 198a | 0.312 | 0.020 | 0.065 | 69.4/69.2 | 91.9/90.4 | 25.4/24.4 | 0.83/0.85 |
| CR057 mitochondrial carrier protein (m) | 350a | 0.315 | 0.011 | 0.036 | 64.9/63.6 | 91.4/87.7 | 27.8/29.2 | 0.80/0.77 | |
| Glyceraldehyde-3-phosphate dehydrogenase | gapC | 195 | 0.340 | 0.014 | 0.040 | 66.3/66.0 | 99.0/98.0 | 23.0/23.7 | 0.88/0.87 |
| ADP/ATP translocator (m) | Ant | 307a | 0.367 | 0.004 | 0.010 | 63.1/63.1 | 90.2/89.6 | 27.5/26.5 | 0.83/0.81 |
| Glycine decarboxylase complex H (m) | GcvH | 158a | 0.367 | 0.013 | 0.036 | 62.9/63.3 | 84.2/85.4 | 32.3/31.3 | 0.71/0.74 |
| NADH:ubiquinone oxidoreductase 13-kDa-like subunit (m) | 153a | 0.370 | 0.016 | 0.044 | 63.4/61.7 | 83.7/78.4 | 43.1/44.8 | 0.61/0.61 | |
| Photosystem I polypeptide 28 (p) | PsaH | 129a | 0.376 | 0.016 | 0.043 | 65.6/66.9 | 84.5/86.8 | 27.8/28.5 | 0.81/0.79 |
| Oxygen-evolving complex/thylakoid Lumenal 25.6-kDa (p) | PsbP | 204 | 0.395 | 0.014 | 0.036 | 64.9/65.4 | 85.8/86.3 | 34.5/31.6 | 0.65/0.69 |
| Inorganic pyrophosphatase (p) | PpaI | 207 | 0.398 | 0.013 | 0.034 | 66.3/66.2 | 90.8/89.8 | 26.8/28.0 | 0.81/0.80 |
| Calmodulin | 162a | 0.398 | 0.002 | 0.006 | 60.5/60.5 | 87.7/87.0 | 36.2/35.6 | 0.71/0.73 | |
| Mago nashi-like protein | 147a | 0.455 | 0.005 | 0.012 | 56.5/55.1 | 85.0/82.3 | 34.2/38.9 | 0.68/0.66 | |
| Carotenoid binding protein (p) | Cbr | 161a | 0.508 | 0.022 | 0.042 | 64.6/64.6 | 82.0/81.4 | 33.0/32.2 | 0.62/0.62 |
| Nucleolar RNA-binding Nop10p-like | 63a | 0.513 | 0.027 | 0.052 | 59.8/60.8 | 76.2/81.0 | 44.5/36.3 | 0.55/0.54 | |
| Minus dominance mating type | mid | 143a | 0.520 | 0.113 | 0.217 | 50.0/48.7 | 57.0/55.4 | 61.0/60.0 | 0.36/0.38 |
| Microtubule-associated protein putative | 132a | 0.540 | 0.032 | 0.059 | 58.1/56.6 | 81.8/77.2 | 45.6/49.6 | 0.56/0.57 | |
| Succinoglycan biosynthesis protein related (m) | 200 | 0.549 | 0.054 | 0.099 | 67.5/64.2 | 84.2/76.6 | 40.0/42.0 | 0.56/0.51 | |
| Hydroxyproline-rich glycoprotein VSP-3 | vsp-3 | 259 | 0.575 | 0.040 | 0.070 | 63.6/63.1 | 83.4/83.0 | 36.3/37.9 | 0.58/0.56 |
| Apospory-associated C protein | apoC | 310 | 0.579 | 0.015 | 0.025 | 67.0/66.3 | 95.5/93.5 | 23.5/29.9 | 0.84/0.81 |
| Peptidylprolyl isomerase/cyclophilin-like | 199a | 0.590 | 0.021 | 0.036 | 68.5/68.2 | 95.0/93.5 | 25.8/25.2 | 0.81/0.84 | |
| PR46b protein | pr46b | 265a | 0.624 | 0.053 | 0.085 | 62.0/65.2 | 70.6/79.2 | 46.9/40.5 | 0.45/0.52 |
| Expressed protein | 107a | 0.647 | 0.035 | 0.055 | 68.5/66.7 | 91.6/86.9 | 30.2/28.7 | 0.67/0.72 | |
| S-adenosyl-l-methionine synthetase | samS | 241 | 0.649 | 0.011 | 0.017 | 65.0/63.3 | 94.2/90.5 | 23.8/26.4 | 0.89/0.84 |
| Pyruvate dehydrogenase kinase-like (m) | Pdk | 487a | 0.695 | 0.032 | 0.046 | 66.7/65.0 | 89.7/84.4 | 32.1/34.5 | 0.60/0.57 |
| Mating-type protein Mat3p/retinoblastoma-like protein | mat3 | 1194a | 0.713 | 0.061 | 0.085 | 67.9/68.0 | 85.0/84.8 | 36.2/35.9 | 0.54/0.54 |
| Alternative oxidase 1 (m) | Aox1 | 260 | 0.734 | 0.017 | 0.023 | 69.9/68.1 | 96.9/92.7 | 26.1/31.0 | 0.74/0.67 |
| Phosphoenolpyruvate carboxykinase related | pck | 217 | 0.793 | 0.020 | 0.025 | 65.0/63.9 | 91.2/86.6 | 30.0/31.0 | 0.76/0.72 |
| Extracellular polypeptide Ecp88 | 229 | 0.805 | 0.033 | 0.041 | 65.6/65.5 | 91.3/90.8 | 25.6/28.4 | 0.74/0.71 | |
| Asparagine synthetase related | 219 | 1.161 | 0.024 | 0.021 | 65.3/64.5 | 87.7/84.9 | 34.1/42.2 | 0.60/0.57 | |
| Striated fiber assemblin | sfa | 203 | 1.194b | 0.008b | 0.007b | 62.9/64.7 | 83.7/88.2 | 38.0/31.8 | 0.64/0.64 |
| Pherophorin I | 231 | 1.682 | 0.120 | 0.071 | 66.1/66.8 | 92.3/92.2 | 27.0/26.0 | 0.85/0.82 | |
| Average | 220 | 0.370 | 0.018 | 0.056 | 64.7/64.4* | 88.2/87.2* | 30.1/30.2 | 0.75/0.75 | |
| Coefficient of variation % | 65 | 79 | 122 | 71 | 5/5 | 7/7 | 23/22 | 15/15 | |
Lc, length of codons analyzed. dS and dN, numbers of synonymous and nonsynonymous substitutions per site, respectively. GC%, percentage of G + C. GC3%, percentage of G + C in the third codon position. m, mitochondrial targeted. p, plastid targeted. * Significantly different at P < 0.01.
Full-length coding sequence.
dS and dN estimated by using the counting method (Yang and Nielsen 2000), because the maximum-likelihood method failed to converge.
The selection of sequences for analysis was not done randomly. Sequence length and quality were considered, and as much as possible we choose genes encoding products that perform diverse cellular functions and are targeted to different cellular compartments. We designated genes as encoding mitochondrial- or plastid-targeted products on the basis of the best BLAST hits for proteins that are known to be targeted to the respective organelles and annotated as such in the GenBank, C. reinhardtii EST, and C. incerta PEP databases. The remaining seven gene sequences were obtained from GenBank or, for C. incerta, by sequencing PCR products. The mean length of the 67 homologous pairs of protein-coding sequences from C. reinhardtii and C. incerta, as aligned codon by codon using Clustal X (Thompson et al. 1997), was 220 codons (maximum, 1194; minimum, 63). The full coding sequence was analyzed in 41 (61%) of the homologous gene pairs examined. The C. reinhardtii and C. incerta intron sequences were aligned using Multalin (Corpet 1988) and then manually adjusted.
Distance estimation:
The number of synonymous substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) in the protein-coding regions were estimated using the maximum-likelihood (ML) method (Goldman and Yang 1994) implemented in the CODEML program of the version 3.14 PAML package (Yang 1997), assuming transition/transversion bias and codon usage bias (F3x4). The number of substitutions per site in the introns (dI) was calculated with the Hasegawa–Kishino–Yano (HKY85) model (Hasegawa et al. 1985) implemented in the BASEML program, also part of the PAML package, assuming transition/tranversion bias and nonuniform base composition.
Codon usage bias:
CODONS (Lloyd and Sharp 1992), MEGA 2.1 (Kumar et al. 2001), and DAMBE (Xia and Xie 2001) software packages were used to compute the Nc (Wright 1990), base composition, RSCU values, and the CAI (Sharp and Li 1987b). In calculating the CAI, we used the specific set of optimal codons previously defined by Naya et al. (2001), which was based on the codon usage frequencies of C. reinhardtii highly expressed genes.
Statistical methods:
The paired t-tests and the calculation of Pearson correlation coefficients were performed using MINITAB, release 14.12.0. The likelihood-ratio test was used as described in the PAML manual.
Nucleotide sequence accession numbers:
Sixty-one C. incerta unique sequences (i.e., clusters or singletons) have been deposited in GenBank under accession nos. DQ122864–DQ122923 and DQ222936. The full C. incerta EST data set (5124 entries) is available at http://amoebidia.bcm.umontreal.ca/public/pepdb/agrm.php/. Partial genomic sequences of C. incerta cblP, GapA, sfa, Tpx, and ytpC1 have been deposited in GenBank under accession nos. DQ122924–DQ122927 and DQ222937.
RESULTS
Comparison of codon usage in C. reinhardtii and C. incerta:
As shown in Table 1, measures of compositional (GC%, GC3%) and especially codon biases (Nc and CAI) vary considerably among genes in C. incerta, and in pairwise comparisons with C. reinhardtii orthologs, the values of these parameters correlate strongly (r > 0.9). Moreover, the averages of these parameters are almost identical between the two species although the small difference in the GC% averages is statistically significant and the same is true for the GC3% averages (Table 1). On the basis of these data, it is not surprising that RSCU values of concatenated C. reinhardtii and C. incerta gene sequences are highly correlated (r = 0.99) (supplemental Table S2 at http://www.genetics.org/supplemental/).
Nc in C. incerta correlates strongly with the level of gene expression as indicated by CAI (r = −0.89, P < 0.001) and with C3 composition (r = −0.76, P < 0.001), but does not correlate significantly with G3 composition (r = 0.07, P = 0.55) (supplemental Figure S1 at http://www.genetics.org/supplemental/). A significant correlation between Nc and the level of gene expression estimated by the number of EST matches of each sequence was previously reported in C. reinhardtii (Naya et al. 2001). We also report a strong positive correlation between CAI and the C content at fourfold degenerate sites in both C. reinhardtii (r = 0.82, P < 0.001) and C. incerta (r = 0.85, P < 0.001) (supplemental Figure S2 at http://www.genetics.org/supplemental/). This finding supports the view that the frequency of C3 increases with the level of gene expression in both taxa and is consistent with the observation of Naya et al. (2001) that highly expressed genes in C. reinhardtii are C3 rich.
Rate of nucleotide substitution among genes:
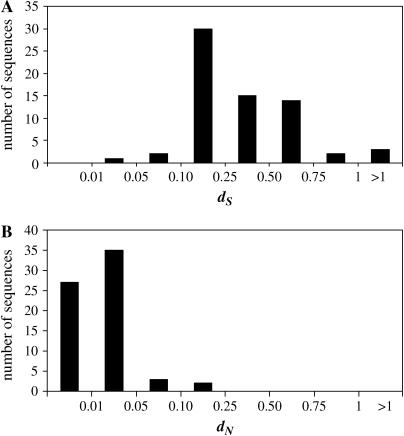
The divergence at synonymous sites, as estimated for individual gene pairs using the ML method (Table 1), varies >65-fold among genes; these dS values range from 0.025 ± 0.008 for cblP to 1.68 ± 0.47 for the pherophorin I gene. Nevertheless, 85% of the genes analyzed exhibit dS values between 0.1 and 0.75 (Figure 1A).
Figure 1.
Distribution of dS (A) and dN (B) for C. incerta and C. reinhardtii protein-coding genes.
Estimates of the nonsynonymous substitution divergence between C. reinhardtii and C. incerta orthologs in our data set range from no divergence for five gene sequences (petF, eIF4A, fla14, Atp9, and yptC1) to values of 0.11 and 0.12 for mid and the gene for pherophorin I, respectively (Table 1). About 90% of the genes show dN values below 0.05 (Figure 1B); however, variation in dN exceeds that of dS by 50% (Table 1).
Finally, we determined dN/dS ratios for the individual genes in our data set (Table 1) and also tested for a correlation between the synonymous and nonsynonymous rates among genes. The dN/dS values vary widely across genes but are consistently <1; the average over all genes is 0.056. At 0.217, the mid gene has the highest dN/dS ratio. Estimates of synonymous and nonsynonymous rates are positively correlated (r = 0.62, P < 0.001).
Correlation between substitution divergence and codon adaptation among genes:
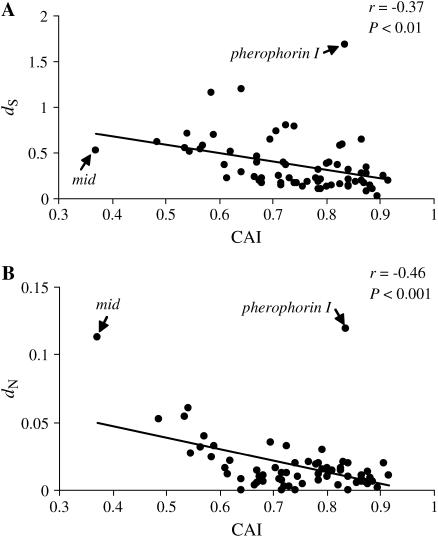
We wanted to determine if the among-gene variation in synonymous and nonsynonymous divergence between C. reinhardtii and C. incerta is correlated with gene expression as indicated by CAI. The analysis reveals a significant negative correlation between dS and CAI (r = −0.37, P < 0.01) (Figure 2A) and the same relationship is also observed between dN and CAI (r = −0.46, P < 0.001) (Figure 2B). mid has a particularly high dN value and pherophorin I is conspicuously high for both dS and dN. Removing these two outliers from the analyses increased the strength of the correlation between dS and CAI (r = −0.50, P < 0.001) and between dN and CAI (r = −0.57, P < 0.001). We found similar correlation coefficients (data not shown) between both dS and dN and the number of ESTs recovered from the C. incerta library (EST abundance data are given in supplemental Table S1 at http://www.genetics.org/supplemental). However, we chose to use CAI rather than EST abundance in our reported analyses because not all genes in our data set were represented in either the C. incerta or the C. reinhardtii library and normalization steps were employed in the preparation of these libraries. In other systems, moreover, CAI has been shown to be as good as mRNA abundance in predicting protein abundance (Jansen et al. 2003) and the best codon-bias-derived surrogate for gene expression level (Goetz and Fuglsang 2005).
Figure 2.
Relationship of dS (A) and dN (B) with the averaged CAI for C. incerta and C. reinhardtii protein-coding genes.
Comparison of intron and exon substitution rate estimates in genes with low- and high-CAI values:
We analyzed concatenated intron and exon sequences from three genes (sfa, mid, and Pdk) with CAI values between 0.36 and 0.64 and three genes (cblP, GapA, and Tpx) with CAI values between 0.81 and 0.90 (the six genes are listed in Table 2; intron data are given in supplemental Table S3 at http://www.genetics.org/supplemental/). Likelihood-ratio tests indicated homogenous rates among sites within introns from the lowly expressed genes (low CAI) and also from the highly expressed genes (high CAI) (supplemental Table S4 at http://www.genetics.org/supplemental/). Next, we found that the difference in substitution rates between the two groups of introns is statistically significant, as the hypothesis of equal substitution rates between the two partitions is rejected by the likelihood-ratio test (2Δℓ = 20.32, d.f. = 1, P < 0.001). The substitution rate in introns of the low-CAI genes is ∼1.3 times higher than that in introns of the high-CAI genes (Table 2). In contrast, when the mean synonymous divergence of the corresponding exons is examined, we find that the synonymous substitution rate in the low-CAI genes is ∼5 times higher than the rate in high-CAI genes (Table 2).
TABLE 2.
Evolutionary divergence (± standard errors) between C. reinhardtii and C. incerta concatenated intron and exon regions of three lowly expressed (low-CAI) and three highly expressed (high-CAI) genes
| Introns
|
Exons
|
||||
|---|---|---|---|---|---|
| Genes | CAI | L | dI | L | dS |
| sfa, mid, Pdk | <0.65 | 3434 | 0.64 ± 0.02 | 2499 | 0.65 ± 0.06 |
| cblP, GapA, Tpx | >0.80 | 1803 | 0.50 ± 0.01 | 2427 | 0.13 ± 0.02 |
L, number of sites analyzed. dI, number of substitutions per site. dS, number of synonymous substitutions per site.
DISCUSSION
Analysis of 67 pairs of orthologous genes revealed similar codon usage and base composition between C. reinhardtii and C. incerta, suggesting the absence of differences in selective or mutational forces acting on codon usage in the two taxa since their divergence.
There is considerable variation in the synonymous and nonsynonymous substitution rates among the C. reinhardtii/C. incerta genes examined even when the extreme values are not considered. The synonymous substitution divergence across the genes studied here is at least as large as the among-gene synonymous rate variation reported in other systems, e.g., bacteria (Sharp and Li 1987a), vertebrates (Bernardi et al. 1993; Wolfe and Sharp 1993; Moriyama and Powell 1997), and land plants (Alvarez-Valin et al. 1999; Kusumi et al. 2002; Tiffin and Hahn 2002; Senchina et al. 2003), although when comparing these values one must consider differences in methods of divergence estimation, sample size, and gene sets, all of which can affect the level of variation in the synonymous divergence among genes.
Evidence for a negative correlation between the estimated synonymous divergence among genes and gene expression level, as supported here for C. reinhardtii/C. incerta, has also been reported for bacteria and yeast (Sharp and Li 1987a; Sharp 1991; Pal et al. 2001; Smith and Eyre-Walker 2001; Rispe et al. 2004; Hirsh et al. 2005) but contrasts with reports on mammals and Arabidopsis where synonymous substitution rate and level of gene expression seem uncorrelated (Duret and Mouchiroud 2000; Wright et al. 2002, 2004). At least two causes for the negative correlation between synonymous substitution rate and expression level of the Chlamydomonas genes can be invoked: (i) synonymous substitution rate is reduced by translational selection, which increases with gene expression; and (ii) highly expressed genes acquire fewer mutations because of transcription-mediated repair processes (e.g., Berg and Martelius 1995; Eyre-Walker and Bulmer 1995; see also Sullivan 1995 for a review). One expects that transcription-coupled mutation-repair processes would affect introns and flanking exons equally. Therefore, if such processes represent the major cause of the dS depression in the exons of high-CAI genes (e.g., cblP, GapA, and Tpx) relative to the exons of low-CAI genes (e.g., sfa, mid, and Pdk), a nearly similar drop in divergence in introns of the former gene set compared to the latter gene set would be expected. Alternatively, if translational selection is the major evolutionary force responsible for the lower dS in the exons of high-CAI genes compared to the exons of low-CAI genes, one might expect equal levels of divergence between the introns from the two sets of genes. Our results fall between these two extremes. There is significantly lower divergence in the introns of the high-CAI genes relative to the low-CAI genes examined. This difference, however, is about four times less than the drop in the synonymous substitution divergence in the exons of the two gene categories. These results, therefore, support translational selection over mutation as the more important evolutionary force underlying the described negative correlation between dS and CAI in the Chlamydomonas taxa. The lower intron evolutionary rate in the high-CAI genes compared to the low-CAI ones could result from transcription-mediated repair processes, although these results are also consistent with enhanced selective constraints on the sequence of introns in highly expressed genes. Nevertheless, in spite of the fact that there are reports showing that spliceosomal introns are subject to constraints in sequence evolution, especially the first introns and the sites flanking intron/exon boundries (e.g., Chamary and Hurst 2004), there is no report indicating that these constraints differ among highly and lowly expressed genes.
The among-gene heterogeneity in the nonsynonymous substitution rates between C. reinhardtii and C. incerta is considerable and shows more variation than the synonymous substitution rates, which is in agreement with the expected variations in functional constraints on the amino acid sequence in different proteins and with reports on other biological groups (Li and Graur 1991; Bernardi et al. 1993; Wolfe and Sharp 1993; Tiffin and Hahn 2002). The negative correlation between the rate of nonsynonymous substitutions and the level of gene expression found in a number of systems (Duret and Mouchiroud 2000; Pal et al. 2001; Wright et al. 2002, 2004; Marais et al. 2004; Rispe et al. 2004; Rocha and Danchin 2004; Subramanian and Kumar 2004; Lemos et al. 2005) has also been observed here in C. reinhardtii/C. incerta. Different models have been proposed to explain this connection. In the translational selection model, highly expressed genes are proposed to be under purifying selection against nonsynonymous changes that may be neutral with respect to protein function but suboptimal with respect to translational efficiency (Akashi 2001, 2003). Other models propose that genes that are broadly expressed across tissues in multicellular eukaryotes, which are also the most highly expressed, are under enhanced functional constraints because their products must function in a greater number of cellular environments (Hastings 1996) or because nonsynonymous changes in these genes affect a greater number of tissues and therefore have a greater impact on fitness (Duret and Mouchiroud 2000). In Chlamydomonas, the relationship between the rate of nonsynonymous substitution and the level of gene expression might be better explained by the translational selection model because there is evidence for translational selection in C. reinhardtii (Naya et al. 2001) and Chlamydomonas taxa are unicellular organisms. Consistent with this idea we found (i) a strong negative correlation between Nc and the level of gene expression in C. incerta, (ii) a significant negative correlation between the rate of synonymous substitution in C. reinhardtii/C. incerta and the level of gene expression, and (iii) a significant correlation of both Nc (data not shown) and synonymous substitution rate with the nonsynonymous substitution rate in C. reinhardtii/C. incerta. Nevertheless, some effects on protein functional constraints related to breadth of gene expression might be expected in C. reinhardtii/C. incerta. These taxa undoubtedly exist under diverse environmental conditions and have both asexual and sexual life-cycle phases so that genes may vary in their breadth of expression under different physiological or developmental stages. If the breadth and level of gene expression are correlated in Chlamydomonas, it may prove difficult to separate their effects on protein evolution.
The highest nonsynonymous substitution estimates in our data set come from mid and the pherophorin I gene, which are approximately six times greater than the value averaged over the whole set of genes. mid encodes a minus-dominance protein important in gamete sex-determination in Chlamydomonas (Ferris and Goodenough 1997). Among both unicellular and multicellular eukaryotes there is evidence that sex-related genes evolve significantly more rapidly than genes not directly related to sex functions (e.g., Singh and Kulathinal 2000; Torgerson and Singh 2003; Zhang et al. 2004). In C. reinhardtii and C. incerta, mid was reported previously to evolve rapidly in terms of nonsynonymous substitutions. However, two regions of the predicted protein product were observed to be conspicuously more conserved in amino acid sequence between these species as compared to two other regions of the protein (Ferris et al. 1997). We searched the InterPro Scan database (http://www.ebi.ac.uk/InterProScan) and found that the C-terminal region previously described as conserved between C. reinhardtii and C. incerta indeed contains a domain conserved across the plant lineage (the RWP-RK domain; Pfam accession no. PF02042). According to our estimates, the average nonsynonymous substitution rate for sites in the nonconserved regions of this gene (dN = 0.28 ± 0.06) is ∼2.5 times higher than the rate averaged over all sites in the gene (dN = 0.113 ± 0.02) and ∼8 times higher than those averaged over the sites in the conserved regions of the gene (dN = 0.035 ± 0.013). The high evolutionary rates at the nonsynonymous sites in the nonconserved regions are consistent with a relaxation of functional constraint or, as proposed by Ferris et al. (1997), positive selection, while the conserved domains are probably under purifying selection. Although the different domains experience different rates at nonsynonymous sites, they have rather similar synonymous substitution rates, base composition, and codon bias.
The pherophorin I gene belongs to a multigene family described so far in Volvox carteri (Godl et al. 1995; Hallmann 2003) and C. reinhardtii (Nedelcu 2005). In V. carteri, which is a colonial close relative of C. reinhardtii/C. incerta, pherophorins are major constituents of the extracellular matrix and are structurally related to the sex-inducing pheromone (reviewed by Hallmann 2003). Divergence at both synonymous and nonsynonymous sites in the pherophorin I gene is the highest in our data set. The points representing the pherophorin gene in both the dS vs. CAI and dN vs. CAI plots lie conspicuously well above the regression lines, suggesting that the expression level had little cause for the high evolutionary rates of this gene. Relaxed purifying selection or positive selection on this gene cannot be invoked to explain the high dN values unless a substantial overestimation of dS is assumed, as the dN/dS ratio is not exceptionally high. Alternatively, these pherophorin I sequences may have existed as ancient alleles or paralogs prior to the C. incerta and C. reinhardtii speciation event and therefore had more time to diverge. In this connection, the pherophorin I genes used in this study are the most closely related pherophorin-like genes currently in the databases of the two taxa on the basis of both phylogenetic affiliation (data not shown) and sequence similarity (the BLAST E-value was at least 100 orders of magnitude smaller for this pair of genes than for other hits).
C. reinhardtii is unquestionably the premier unicellular model organism among green plants (Harris 2001; Gutman and Niyogi 2004). Yet the exciting potential of this system for molecular evolutionary analysis was hindered by the scarcity of appropriate comparative sequence data. The placement of our newly generated cDNA sequence data for C. incerta in a comparative framework with the existing gene sequences from C. reinhardtii has opened the opportunity to address fundamental questions about the relative roles of translational selection and transcription-coupled repair processes in the nuclear compartment of this model group of green algae. Although this study represents only the first step in addressing these and related questions, the results should prove valuable in guiding future comparative and experimental work. For example, it is important that we test our findings by employing other measures of gene expression such as identifying genes richest in codons having the most abundant cognate tRNAs and by measuring transcript abundance using microarray analysis. Finally, with a larger sample and improved annotation of orthologous genes from these taxa it should be possible to investigate attributes such as gene age, gene length, and codon usage bias along the translational gradient that have been shown in other systems to have fine-scale effects in shaping the rates and patterns of nucleotide sequence evolution.
Acknowledgments
We thank Aurora Nedelcu, University of New Brunswick at Fredericton, for helpful discussions on pherophorins and for providing alignments of C. reinhardtii pherophorin-like sequences. This work was supported by Natural Sciences and Engineering Research Council of Canada grants to R.W.L and J.P.B. and is part of the Protist EST Program (PEP) funded by Genome Canada and the Atlantic Canada Opportunities Agency (Atlantic Innovation Fund). C.E.P. received scholarships from Dalhousie University and the Patrick F. Lett Fund and T.B. was the recipient of a PEP postdoctoral fellowship.
References
- Akashi, H., 2001. Gene expression and molecular evolution. Curr. Opin. Genet. Dev. 11: 660–666. [DOI] [PubMed] [Google Scholar]
- Akashi, H., 2003. Translational selection and yeast proteome evolution. Genetics 164: 1291–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H., and A. Eyre-Walker, 1998. Translational selection and molecular evolution. Curr. Opin. Genet. Dev. 8: 688–693. [DOI] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Alvarez-Valin, F., K. Jabbari, N. Carels and G. Bernardi, 1999. Synonymous and nonsynonymous substitutions in genes from Gramineae: intragenic correlations. J. Mol. Evol. 49: 330–342. [DOI] [PubMed] [Google Scholar]
- Berg, O. G., and M. Martelius, 1995. Synonymous substitution-rate constants in Escherichia coli and Salmonella typhimurium and their relationship to gene expression and selection pressure. J. Mol. Evol. 41: 449–456. [DOI] [PubMed] [Google Scholar]
- Bernardi, G., D. Mouchiroud and C. Gautier, 1993. Silent substitutions in mammalian genomes and their evolutionary implications. J. Mol. Evol. 37: 583–589. [DOI] [PubMed] [Google Scholar]
- Bierne, N., and A. Eyre-Walker, 2003. The problem of counting sites in the estimation of the synonymous and nonsynonymous substitution rates: implications for the correlation between the synonymous substitution rate and codon usage bias. Genetics 165: 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamary, J. V., and L. D. Hurst, 2004. Similar rates but different modes of sequence evolution in introns and at exonic silent sites in rodents: evidence for selectively driven codon usage. Mol. Biol. Evol. 21: 1014–1023. [DOI] [PubMed] [Google Scholar]
- Chiapello, H., F. Lisacek, M. Caboche and A. Henaut, 1998. Codon usage and gene function are related in sequences of Arabidopsis thaliana. Gene 209: GC1–GC38. [DOI] [PubMed] [Google Scholar]
- Coleman, A. W., and J. C. Mai, 1997. Ribosomal DNA ITS-1 and ITS-2 sequence comparisons as a tool for predicting genetic relatedness. J. Mol. Evol. 45: 168–177. [DOI] [PubMed] [Google Scholar]
- Corpet, F., 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16: 10881–10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, K. A., J. P. Bielawski and Z. Yang, 2001. Substitution rates in Drosophila nuclear genes: implications for translational selection. Genetics 157: 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L., and D. Mouchiroud, 1999. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA 96: 4482–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret, L., and D. Mouchiroud, 2000. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol. Biol. Evol. 17: 68–74. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker, A., and M. Bulmer, 1995. Synonymous substitution rates in enterobacteria. Genetics 140: 1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P. J., and U. W. Goodenough, 1997. Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris, P. J., C. Pavlovic, S. Fabry and U. W. Goodenough, 1997. Rapid evolution of sex-related genes in Chlamydomonas. Proc. Natl. Acad. Sci. USA 94: 8634–8639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godl, K., A. Hallmann, A. Rappel and M. Sumper, 1995. Pherophorins: a family of extracellular matrix glycoproteins from Volvox structurally related to the sex-inducing pheromone. Planta 196: 781–787. [PubMed] [Google Scholar]
- Goetz, R. M., and A. Fuglsang, 2005. Correlation of codon bias measures with mRNA levels: analysis of transcriptome data from Escherichia coli. Biochem. Biophys. Res. Commun. 327: 4–7. [DOI] [PubMed] [Google Scholar]
- Goldman, N., and Z. Yang, 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11: 725–736. [DOI] [PubMed] [Google Scholar]
- Gouy, M., and C. Gautier, 1982. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 10: 7055–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman, A. R., E. E. Harris, C. Hauser, P. A. Lefebvre, D. Martinez et al., 2003. Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell 2: 1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman, B. L., and K. K. Niyogi, 2004. Chlamydomonas and Arabidopsis. A dynamic duo. Plant Physiol. 135: 607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallmann, A., 2003. Extracellular matrix and sex-inducing pheromone in Volvox. Int. Rev. Cytol. 227: 131–182. [DOI] [PubMed] [Google Scholar]
- Harris, E. H., 1989. The Chlamydomonas Sourcebook. Academic Press, San Diego.
- Harris, E. H., 2001. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 52: 363–406. [DOI] [PubMed] [Google Scholar]
- Hasegawa, M., H. Kishino and T. Yano, 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22: 160–174. [DOI] [PubMed] [Google Scholar]
- Hastings, K. E., 1996. Strong evolutionary conservation of broadly expressed protein isoforms in the troponin I gene family and other vertebrate gene families. J. Mol. Evol. 42: 631–640. [DOI] [PubMed] [Google Scholar]
- Hirsh, A. E., H. B. Fraser and D. P. Wall, 2005. Adjusting for selection on synonymous sites in estimates of evolutionary distance. Mol. Biol. Evol. 22: 174–177. [DOI] [PubMed] [Google Scholar]
- Jansen, R., H. J. Bussemaker and M. Gerstein, 2003. Revisiting the codon adaptation index from a whole-genome perspective: analyzing the relationship between gene expression and codon occurrence in yeast using a variety of models. Nucleic Acids Res. 31: 2242–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski, L. B., M. W. Gray, B. F. Lang and G. Burger, 2005. AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics 6: 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., K. Tamura, I. B. Jakobsen and M. Nei, 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17: 1244–1245. [DOI] [PubMed] [Google Scholar]
- Kusumi, J., Y. Tsumura, H. Yoshimaru and H. Tachida, 2002. Molecular evolution of nuclear genes in Cupressacea, a group of conifer trees. Mol. Biol. Evol. 19: 736–747. [DOI] [PubMed] [Google Scholar]
- LeDizet, M., and G. Piperno, 1995. The light chain p28 associates with a subset of inner dynein arm heavy chains in Chlamydomonas axonemes. Mol. Biol. Cell 6: 697–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos, B., B. R. Bettencourt, C. D. Meiklejohn and D. L. Hartl, 2005. Evolution of proteins and gene expression levels are coupled in Drosophila and are independently associated with mRNA abundance, protein length, and number of protein-protein interactions. Mol. Biol. Evol. 22: 1345–1354. [DOI] [PubMed] [Google Scholar]
- Li, W.-H., and D. Graur, 1991. Fundamentals of Molecular Evolution. Sinauer Associates, Sunderland, MA.
- Liss, M., D. L. Kirk, K. Beyser and S. Fabry, 1997. Intron sequences provide a tool for high-resolution phylogenetic analysis of volvocine algae. Curr. Genet. 31: 214–227. [DOI] [PubMed] [Google Scholar]
- Lloyd, A. T., and P. M. Sharp, 1992. CODONS: a microcomputer program for codon usage analysis. J. Hered. 83: 239–240. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., E. F. Fritsch and J. Sambrook, 1982. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Marais, G., T. Domazet-Loso, D. Tautz and B. Charlesworth, 2004. Correlated evolution of synonymous and nonsynonymous sites in Drosophila. J. Mol. Evol. 59: 771–779. [DOI] [PubMed] [Google Scholar]
- Moriyama, E. N., and J. R. Powell, 1997. Synonymous substitution rates in Drosophila: mitochondrial versus nuclear genes. J. Mol. Evol. 45: 378–391. [DOI] [PubMed] [Google Scholar]
- Naya, H., H. Romero, N. Carels, A. Zavala and H. Musto, 2001. Translational selection shapes codon usage in the GC-rich genome of Chlamydomonas reinhardtii. FEBS Lett. 501: 127–130. [DOI] [PubMed] [Google Scholar]
- Nedelcu, A. M., 2005. Sex as a response to oxidative stress: stress genes co-opted for sex. Proc. R. Soc. Lond. B Biol. Sci. 272: 1935–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H., 2003. Neutral mutations and neutral substitutions in bacterial genomes. Mol. Biol. Evol. 20: 2091–2096. [DOI] [PubMed] [Google Scholar]
- Pal, C., B. Papp and L. D. Hurst, 2001. Highly expressed genes in yeast evolve slowly. Genetics 158: 927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, J. R., and E. N. Moriyama, 1997. Evolution of codon usage bias in Drosophila. Proc. Natl. Acad. Sci. USA 94: 7784–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold, T., E. H. Harris and A. Coleman, 2005. Portrait of a species: Chlamydomonas reinhardtii. Genetics 170: 1601–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold, T., B. Marin, U. G. Schlösser and M. Melkonian, 2001. Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta). I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist 152: 265–300. [DOI] [PubMed] [Google Scholar]
- Rispe, C., F. Delmotte, R. C. van Ham and A. Moya, 2004. Mutational and selective pressures on codon and amino acid usage in Buchnera, endosymbiotic bacteria of aphids. Genome Res. 14: 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha, E. P., and A. Danchin, 2004. An analysis of determinants of amino acids substitution rates in bacterial proteins. Mol. Biol. Evol. 21: 108–116. [DOI] [PubMed] [Google Scholar]
- Schlösser, U. G., H. Sachs and D. G. Robinson, 1976. Isolation of protoplasts by means of a ‘species-specific’ autolysine in Chlamydomonas. Protoplasma 88: 51–64. [DOI] [PubMed] [Google Scholar]
- Senchina, D. S., I. Alvarez, R. C. Cronn, B. Liu, J. Rong et al., 2003. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 20: 633–643. [DOI] [PubMed] [Google Scholar]
- Sharp, P. M., 1991. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J. Mol. Evol. 33: 23–33. [DOI] [PubMed] [Google Scholar]
- Sharp, P. M., and W. H. Li, 1987. a The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol. Biol. Evol. 4: 222–230. [DOI] [PubMed] [Google Scholar]
- Sharp, P. M., and W. H. Li, 1987. b The codon Adaptation Index: a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15: 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P. M., T. M. Tuohy and K. R. Mosurski, 1986. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 14: 5125–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, R. S., and R. J. Kulathinal, 2000. Sex gene pool evolution and speciation: a new paradigm. Genes Genet. Syst. 75: 119–130. [DOI] [PubMed] [Google Scholar]
- Smith, N. G., and A. Eyre-Walker, 2001. Nucleotide substitution rate estimation in enterobacteria: approximate and maximum-likelihood methods lead to similar conclusions. Mol. Biol. Evol. 18: 2124–2126. [DOI] [PubMed] [Google Scholar]
- Stenico, M., A. T. Lloyd and P. M. Sharp, 1994. Codon usage in Caenorhabditis elegans: delineation of translational selection and mutational biases. Nucleic Acids Res. 22: 2437–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S., and S. Kumar, 2004. Gene expression intensity shapes evolutionary rates of the proteins encoded by the vertebrate genome. Genetics 168: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, D. T., 1995. DNA excision repair and transcription: implications for genome evolution. Curr. Opin. Genet. Dev. 5: 786–791. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffin, P., and M. W. Hahn, 2002. Coding sequence divergence between two closely related plant species: Arabidopsis thaliana and Brassica rapa ssp. pekinensis. J. Mol. Evol. 54: 746–753. [DOI] [PubMed] [Google Scholar]
- Torgerson, D. G., and R. S. Singh, 2003. Sex-linked mammalian sperm proteins evolve faster than autosomal ones. Mol. Biol. Evol. 20: 1705–1709. [DOI] [PubMed] [Google Scholar]
- Wolfe, K. H., and P. M. Sharp, 1993. Mammalian gene evolution: nucleotide sequence divergence between mouse and rat. J. Mol. Evol. 37: 441–456. [DOI] [PubMed] [Google Scholar]
- Wright, F., 1990. The ‘effective number of codons’ used in a gene. Gene 87: 23–29. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., B. Lauga and D. Charlesworth, 2002. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 19: 1407–1420. [DOI] [PubMed] [Google Scholar]
- Wright, S. I., C. B. Yau, M. Looseley and B. C. Meyers, 2004. Effects of gene expression on molecular evolution in Arabidopsis thaliana and Arabidopsis lyrata. Mol. Biol. Evol. 21: 1719–1726. [DOI] [PubMed] [Google Scholar]
- Xia, X., and Z. Xie, 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92: 371–373. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., and R. Nielsen, 2000. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 17: 32–43. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., T. M. Hambuch and J. Parsch, 2004. Molecular evolution of sex-biased genes in Drosophila. Mol. Biol. Evol. 21: 2130–2139. [DOI] [PubMed] [Google Scholar]