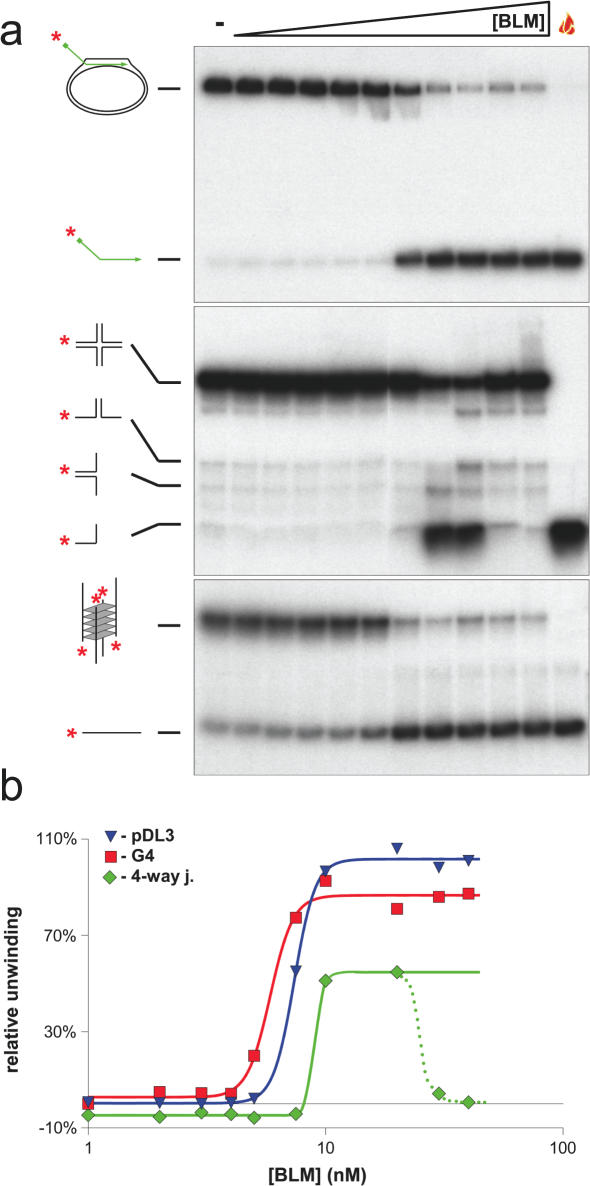
Figure 4.
Comparison of the efficiency of unwinding of different substrates by BLM. BLM enzyme-concentration dependent unwinding of the pDL3 mobile D-loop (upper panel), the X12 four-way junction (middle panel) and G4 (lower panel) substrates. The enzyme-concentration was varied between 1–40 nM as indicated above by the triangle. Reactions lacking BLM are denoted by ‘−’ above. The flame symbol denotes heat-denatured DNA. The substrates and the products of unwinding are shown on the left of each panel. (b) Quantification of the data from (a). Blue triangle, green diamond and red square symbols depict the pDL3, four-way junction and G4 DNA substrates, respectively. Analysis of the concentration-dependence curve indicated that 50% unwinding of pDL3, four-way junction and G4 DNA is achieved at 7.38, 8.97 and 5.84 nM, respectively. At high concentrations of BLM, the strand annealing activity of the enzyme becomes dominant over the unwinding activity, resulting in a diminution in the level of unwound product for the four-way junction substrate [middle panel of (a), green triangles and dotted line of (b)]. The last two concentration points (30 and 40 nM) were, therefore, omitted from the non-linear regression analysis and for determination of the 50% unwinding concentration (see Materials and Methods).