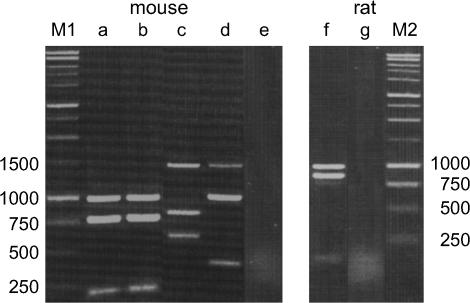
Figure 4.
Restriction enzyme analyses of transcripts of Mkrn1 and its processed pseudogenes in Mus and Rattus. The RT–PCR products were digested by HindIII, EcoRI, or BglII and separated by electrophoresis on a 1.5% agarose gel. (Lanes a and b) In M. m. domesticus and M. spretus (SPRET/Ei), there is a HindIII site in Mkrn1, but not in Mkrn1-p1. (Lane c) In M. caroli, a HindIII site is present in Mkrn1-p1.a, but absent in Mkrn1-p1.b, Mkrn1, and Mkrn1-p2. (Lane d) In M. caroli, an EcoRI site is present in Mkrn1-p1.a/b and Mkrn1, but absent in Mkrn1-p2. (Lane f) In R. norvegicus, Mkrnp1 has a BglII site, but Mkrn1 does not. Lanes e and g represent negative controls in M. caroli and R. norvegicus, respectively. These control experiments were carried out using total RNA to confirm no contamination of genomic DNA in samples. The molecular weight standards are shown on the left- and rightmost lanes (M1 and M2).