TABLE 3.
Molecular markers and map positions
|
D. santomea
|
D. yakuba
|
|||||
|---|---|---|---|---|---|---|
| Marker | Marker name | Cytological location | r | Genetic distance (cM) | r | Genetic distance (cM) |
| Chromosome X | ||||||
| 1 | y | 1A5 | 0.0432 | 0.0 | 0.0245 | 0.0 |
| 2 | per | 3B1–2 | 0.1572 | 4.5 | 0.1468 | 2.5 |
| 3 | sog | 13E1 | 0.1883 | 23.4 | 0.1364 | 19.9 |
| 4 | v | 9F11 | 0.3005 | 47.0 | 0.2690 | 35.8 |
| 5 | rux | 5D2 | 0.0570 | 93.0 | 0.0913 | 74.4 |
| 6 | f | 15F4–7 | 0.0777 | 99.0 | 0.1110 | 84.5 |
| 7 | bnb | 17D6 | 0.0363 | 107.5 | 0.0329 | 97.2 |
| 8 | Hex-A | 8E10 | 0.0760 | 111.2 | 0.0593 | 100.6 |
| 9 | AnnX | 19C1 | 0.0501 | 119.5 | 0.0254 | 106.9 |
| 10 | su(f) | 20E | 0.0000 | 124.7 | 0.0000 | 109.5 |
| Chromosome 2 | ||||||
| 11 | l(2)gl | 21A5 | 0.0639 | 0.0 | 0.0865 | 0.0 |
| 12 | Rad1 | 23A1 | 0.1054 | 6.8 | 0.0922 | 9.5 |
| 13 | RpL27A | 24F3 | 0.1364 | 18.7 | 0.1665 | 19.7 |
| 14 | salr | 32E4–F1 | 0.1002 | 34.6 | 0.1001 | 39.9 |
| 15 | Rep4 | 34B4 | 0.2073 | 45.8 | 0.2023 | 51.2 |
| 16 | His3 | 39D3–E1 | 0.0345 | 72.6 | 0.0132 | 77.1 |
| 17 | barr | 38B1–2 | 0.1140 | 76.1 | 0.1477 | 78.4 |
| 18 | Sara | 57E6 | 0.2694 | 89.1 | 0.2653 | 96.0 |
| 19 | Kr | 60F5 | 0.0000 | 127.8 | 0.0000 | 133.8 |
| Chromosome 3 | ||||||
| 20 | Lsp1γ | 61A6 | 0.2729 | 0.0 | 0.1966 | 0.0 |
| 21 | Dib | 64A5 | 0.2712 | 39.5 | 0.1345 | 25.0 |
| 22 | sfl | 65B3–4 | 0.2314 | 78.5 | 0.2700 | 40.6 |
| 23 | Est-6 | 69A1 | 0.3057 | 109.6 | 0.2493 | 79.5 |
| 24 | Ssl1 | 80B2 | 0.2211 | 156.9 | 0.1637 | 114.0 |
| 25 | ry | 87D9 | 0.2159 | 186.1 | 0.1797 | 133.8 |
| 26 | Rpn5 | 83C4 | 0.0967 | 214.3 | 0.1176 | 156.1 |
| 27 | AP-50 | 94A15–16 | 0.2297 | 225.1 | 0.1844 | 169.5 |
| 29 | Mlc1 | 98A14–15 | 0.1036 | 255.8 | 0.1072 | 192.5 |
| 30 | ymp | 96E | 0.2107 | 267.4 | 0.1110 | 204.6 |
| 28 | janB | 99D3 | 0.2055 | 294.8 | 0.1326 | 217.1 |
| 31 | krz | 100E3 | 0.0000 | 321.3 | 0.0000 | 232.5 |
| Chromosome 4 | ||||||
| 32 | ci | 102A1–3 | 0.0000 | NA | 0.0000 | NA |
r is the recombination rate between two adjacent markers. The genetic distance d was inferred from r using the Haldane map function, 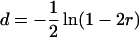 . Cytological locations are given on the basis of D. melanogaster cytology (Lemeunier and Ashburner 1976).
. Cytological locations are given on the basis of D. melanogaster cytology (Lemeunier and Ashburner 1976).
