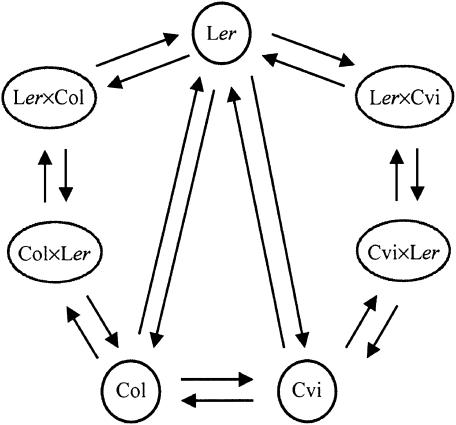
Figure 1.
The loop design, consisting of nine replicated dye-swap experiments in which two independent pools of first leaf pair samples of the seven genotypes were compared in an unequal treatment replication structure, on a total of 18 cDNA microarrays. The seven genotypes hybridized on the microarrays were the three homozygous accessions Col, Ler, and Cvi and the two pairs of reciprocal F1 hybrids obtained by crossing Col and Ler and Ler and Cvi. These microarrays had duplicated sets of 6008 Arabidopsis cDNA clones (of which 4876 were used in the analysis), representing almost one-quarter of the Arabidopsis gene repertoire, and 520 control cDNAs. Each microarray is represented by an arrow and connects the two sampled genotypes hybridized to it. The samples at the tail and head of each arrow are labeled with Cy3 and Cy5, respectively. In this arrangement, each sample is labeled equally often with Cy5 and with Cy3, ensuring that expression levels are free from biases due to dye effects. The pairings of pools sampled at days 21 and 24 after germination are represented by the loop in clockwise and anticlockwise directions, respectively.