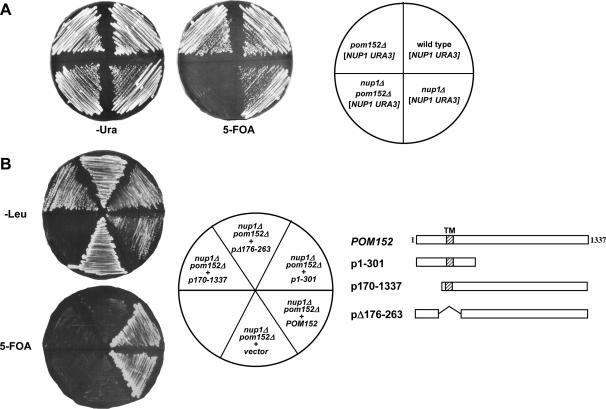
Figure 5.
nup1Δ growth defects are suppressed by pom152 alleles lacking glycosylation. (A) Wild-type (S288C), pom152Δ (KBY1046), nup1Δ (KBY1048), and pom152Δ nup1Δ (KBY1047) cells containing pLDB59 (CEN URA3 NUP1) were streaked on SD −Ura and 5-FOA media, incubated at 25° for 4 days, and scored for growth. (B) nup1Δ pom152Δ (KBY1047) cells containing a CEN URA3 NUP1 plasmid were transformed with plasmids expressing full-length POM152, deletions of N-terminal (p170-1337), transmembrane (pΔ176-263), and lumenal (p1-301) Pom152 domains (Tcheperegine et al. 1999), and empty vector (pRS315). Transformants were streaked on SD −Leu and 5-FOA and incubated at 24°. Streaks were scored for growth after 3 days on SD −Leu and after 5 days on 5-FOA. (C) POM152, pom152ΔGlyc (KBB382), pom152ΔC (p1-301), and empty vector (pRS315) were transformed into nup1Δ pom152Δ (KBY1047) cells containing a CEN URA3 NUP1 plasmid. Transformants were streaked to SD −Leu and 5-FOA and incubated at 24° and at 37°.