Figure 1.
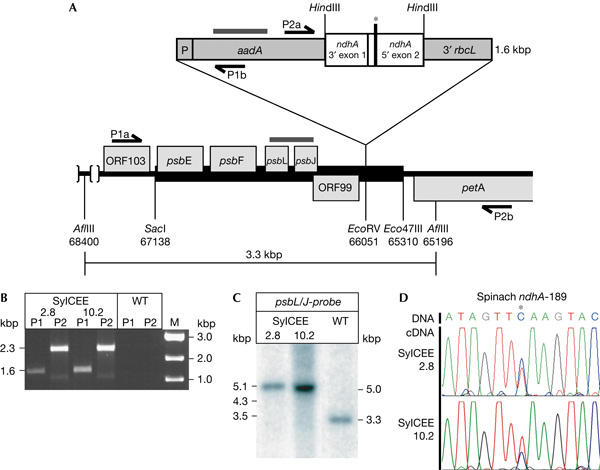
Introduction of a recombinant spinach ndhA construct into the Nicotiana sylvestris plastid chromosome. (A) Schematic representation of parts of the plasmid vector pCEE, as described by Schmitz-Linneweber et al (2001). The thick black bar represents the tobacco plastid DNA fragment used for targeting the recombinant cassette to the petA–psbJ intergenic spacer. The grey bars indicate regions corresponding to probes used in Southern and RNA gel blot analysis. P, 16S ribosomal DNA promoter; 3′rbcL, 3′ stabilizing element. Arrows indicate primers. The asterisk denotes the position of the editing site ndhA-189. Numbers refer to positions in the tobacco plastid chromosome (accession no. Z00044). (B) PCR analysis of correct integration of the transgene into the N. sylvestris plastid chromosome in the two spectinomycine-resistant subclones SylCEE-2.8 and SylCEE-10.2 recovered after transformation. PCR product P1, 1,624 bp; P2, 2,320 bp (see (A) for positions of primers). M, molecular weight marker. (C) Southern analysis of AflIII-digested total genomic DNA (5 μg) of transplastomic lines SylCEE-2.8 and SylCEE-10.2. The probe covers the psbL/J genes (see (A)). Signals obtained correspond to calculated fragment length (indicated on the left; also see (A)). No signal corresponding to wild-type plastid DNA was detected in transplastomic plants, suggesting that they are homoplastomic for the transgene. (D) Complementary DNA sequence analysis of the introduced ndhA editing site in transplastomic lines SylCEE-2.8 and SylCEE-10.2. An excerpt of the corresponding chromatograms is shown with the edited nucleotide in the centre marked by an asterisk. A C-peak corresponding to unedited messages and a T-peak corresponding to edited messages were found in the plant lines analysed (top: corresponding DNA sequence).
