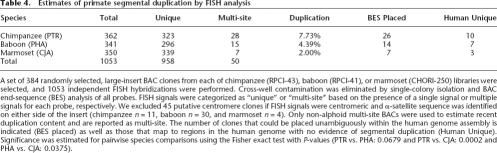
Table 4.
Estimates o primate segmental duplication by FISH analysis
A set of 384 randomly selected, large-insert BAC clones from each of chimpanzee (RPCI-43), baboon (RPCI-41), or marmoset (CHORI-250) libraries were selected, and 1053 independent FISH hybridizations were performed. Cross-well contamination was eliminated by single-colony isolation and BAC end-sequence (BES) analysis of all probes. FISH signals were categorized as “unique” or “multi-site” based on the presence of a single signal or multiple signals for each probe, respectively. We excluded 45 putative centromere clones if FISH signals were centromeric and α-satellite sequence was identified on either side of the insert (chimpanzee n = 11, baboon n = 30, and marmoset n = 4). Only non-alphoid multi-site BACs were used to estimate recent duplication content and are reported as multi-site. The number of clones that could be placed unambiguously within the human genome assembly is indicated (BES placed) as well as those that map to regions in the human genome with no evidence of segmental duplication (Human Unique). Significance was estimated for pairwise species comparisons using the Fisher exact test with P-values (PTR vs. PHA: 0.0679 and PTR vs. CJA: 0.0002 and PHA vs. CJA: 0.0375).