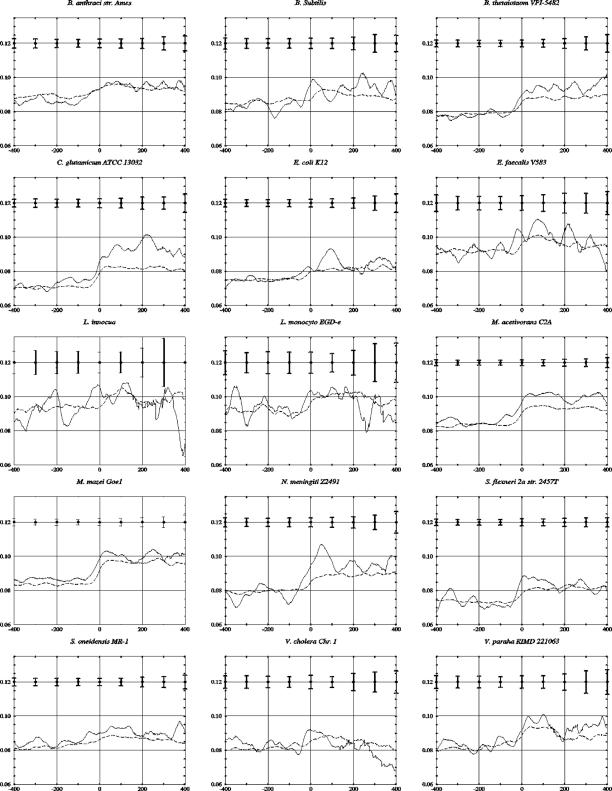
Figure 7.
Curvature distributions in the neighborhood of the end of translation in convergent genes. From 15 selected genomes, which had relatively high curvature excess (DIE), only convergent genes were extracted. The program CURVATURE with a window size of 125 nt was used to predict curvature distributions. Only genes with intergenic region longer than 50 nt were processed. Bacillus anthraci str. Ames (781 genes), B.subtilis (543 genes), B.thetaiotaom VPI-5482 (578 genes), C.glutamicum ATCC 13032 (488 genes), E.coli K12 (632 genes), Enterococcus faecalis V583 (327 genes), Listeria innocua (290 genes), Listeria monocyto EGD-e (301 genes), M.acetivorans C2A (755 genes), M.mazei Goe1 (534 genes), N.meningiti Z2491 (277 genes), Shigella flexneri 2a str. 2457T (626 genes), Shewanella oneidensis MR-1 (778 genes), V.cholera chromosome I (781 genes) and V.paraha RIMD 221063 chromosome I (477 genes). DNA curvature calculations for the real and reshuffled convergent genes of every genome were performed as described in the legend to Figure 1.