Abstract
microRNAs are a highly conserved class of noncoding RNAs with important regulatory functions in proliferation, apoptosis, development, and differentiation. To discover novel regulatory pathways during megakaryocytic differentiation, we performed microRNA expression profiling of in vitro-differentiated megakaryocytes derived from CD34+ hematopoietic progenitors. The main finding was down-regulation of miR-10a, miR-126, miR-106, miR-10b, miR-17 and miR-20. Hypothetically, the down-regulation of microRNAs unblocks target genes involved in differentiation. We confirmed in vitro and in vivo that miR-130a targets the transcription factor MAFB, which is involved in the activation of the GPIIB promoter, a key protein for platelet physiology. In addition, we found that miR-10a expression in differentiated megakaryocytes is inverse to that of HOXA1, and we showed that HOXA1 is a direct target of miR-10a. Finally, we compared the microRNA expression of megakaryoblastic leukemic cell lines with that of in vitro differentiated megakaryocytes and CD34+ progenitors. This analysis revealed up-regulation of miR-101, miR-126, miR-99a, miR-135, and miR-20. Our data delineate the expression of microRNAs during megakaryocytopoiesis and suggest a regulatory role of microRNAs in this process by targeting megakaryocytic transcription factors.
Keywords: leukemia, hematopoiesis
MicroRNAs (miRNAs) are a small noncoding family of 19- to 25-nt RNAs that regulate gene expression by targeting mRNAs in a sequence specific manner, inducing translational repression or mRNA degradation, depending on the degree of complementarity between miRNAs and their targets (1, 2). Many miRNAs are conserved in sequence between distantly related organisms, suggesting that these molecules participate in essential processes. Indeed, miRNAs are involved in the regulation of gene expression during development (3), cell proliferation (4), apoptosis (5), glucose metabolism (6), stress resistance (7), and cancer (8–11).
There is also strong evidence that miRNAs play a role in mammalian hematopoiesis. In mice, miR-181, miR-223, and miR-142 are differentially expressed in hematopoietic tissues, and their expression is regulated during hematopoiesis and lineage commitment (12). The ectopic expression of miR-181 in murine hematopoietic progenitor cells led to proliferation in the B cell compartment (12). Systematic miRNA gene profiling in cells of the murine hematopoietic system revealed different miRNA expression patterns in the hematopoietic system compared with neuronal tissues and identified individual miRNA expression changes that occur during cell differentiation (13). A recent study has identified down modulation of miR-221 and miR-222 in human erythropoietic cultures of CD34+ cord blood progenitor cells (14). These miRNAs were found to target the oncogene c-Kit. Further functional studies indicated that the decline of these two miRNAs in erythropoietic cultures unblocks Kit protein production at the translational level leading to expansion of early erythroid cells (14). In line with the hypothesis of miRNAs regulating cell differentiation, miR-223 was found to be a key member of a regulatory circuit involving C/EBPa and NFI-A, which control granulocytic differentiation in all-trans retinoic acid-treated acute promyelocytic leukemic cell lines (15).
miRNAs have also been found deregulated in hematopoietic malignancies. Indeed, the first report linking miRNAs and cancer involved the deletion and down-regulation of the miR-15a and miR-16-1 cluster, located at chromosome 13q14.3, a commonly deleted region in chronic lymphocytic leukemia (8). High expression of miR-155 and host gene BIC also was reported in B cell lymphomas (16). More recently it was shown that the miR-17-92 cluster, which is located in a genomic region of amplification in lymphomas, is overexpressed in human B cell lymphomas and the enforced expression of this cluster acted in concert with c-MYC expression to accelerate tumor development in a mouse B cell lymphoma model (10). These observations indicate that miRNAs are important regulators of hematopoiesis and can be involved in malignant transformation.
Discovering the patterns and sequence of miRNA expression during hematopoietic differentiation may provide insights about the functional roles of these tiny noncoding genes in normal and malignant hematopoiesis.
In the present study, we investigate the miRNA gene expression in human megakaryocyte cultures from bone marrow CD34+ progenitors and in acute megakaryoblastic leukemia cell lines. The results of this analysis indicate that several miRNAs are down-regulated during normal megakaryocytic differentiation. We demonstrate that these miRNAs target genes involved in megakaryocytopoiesis, whereas others are overexpressed in cancer cells.
Results and Discussion
miRNA Expression During in Vitro Megakaryocytic Differentiation of CD34+ Progenitors.
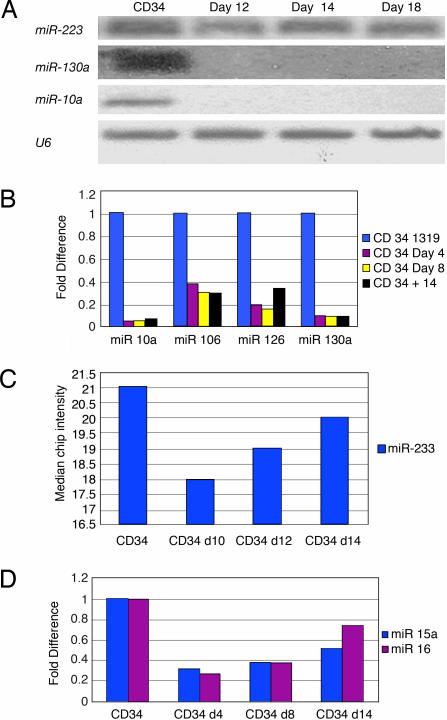
Using a combination of a specific megakaryocytic growth factor (thrombopoietin) and nonspecific cytokines (stem cell factor and IL-3), we were able to generate in vitro pure, abundant megakaryocyte progeny from CD34+ bone marrow progenitors suitable for microarray studies (Fig. 4, which is published as supporting information on the PNAS web site). Total RNA was obtained for miRNA chip analysis from three different CD34 progenitors at baseline and at days 10, 12, 14, and 16 of culture with cytokines. We initially compared the expression of miRNA between the CD34+ progenitors and the pooled CD34+ differentiated megakaryocytes at all points during the differentiation process. We identified 19 miRNAs (Table 1) that are sharply down-regulated during megakaryocytic differentiation. There were no statistically significant miRNAs up-regulated during megakaryocytic differentiation. Using predictive analysis of microarray (PAM) we identified 8 microRNAs that predicted megakaryocytic differentiation with no misclassification error: miR-10a, miR-10b, miR-30c, miR-106, miR-126, miR-130a, miR-132, and miR-143 (Table 3, which is published as supporting information on the PNAS web site). All of these miRNAs, except miR-143, are included in the 17 miRNAs identified by significance analysis of microarray. Northern blots and real-time PCR for several miRNAs confirmed the results obtained by miRNA chip analysis (Fig. 1).
Table 1.
miRNAs down-regulated during in vitro CD34+ megakaryocytic differentiation
| miRNA | Chromosomal location | t test* | Fold change | Putative targets |
|---|---|---|---|---|
| hsa-mir-010a† | 17q21 | −9.10 | 50.00 | HOXA1, HOXA3, HOXD10, CRK, FLT1 |
| hsa-mir-126† | 9q34 | −2.73 | 8.33 | CRK, EVI2, HOXA9, MAFB, CMAF |
| hsa-mir-106† | xq26.2 | −2.63 | 2.86 | TAL1, FLT1, SKI, RUNX1, FOG2, FLI, PDGFRA, CRK |
| hsa-mir-010b† | 2q31 | −2.17 | 11.11 | HOXA1, HOXA3, HOXD10, ETS-1, CRK, FLT1 |
| hsa-mir-130a† | 11q12 | −2.08 | 4.76 | MAFB, MYB, FOG2, CBFB, PDGFRA, SDFR1, CXCL12 |
| hsa-mir-130a-prec† | 11q12 | −2.07 | 7.69 | NA‡ |
| hsa-mir-124a | 8q23 | −1.81 | 2.78 | TAL1, SKI, FLT1, FOG2, ETS-1, CBFB, RAF1, MYB |
| hsa-mir-032-prec | 9q31 | −1.76 | 3.57 | NA‡ |
| hsa-mir-101 | 1p31.3 | −1.75 | 3.33 | TAL1, CXCL12, MEIS1, MEIS2, ETS-1 RUNX1, MYB |
| hsa-mir-30c | 6q13 | −1.71 | 2.56 | CBFB, MAFG, HOXA1, SBF1, NCOR2, ERG |
| hsa-mir-213† | 1q31.3 | −1.69 | 2.38 | MAX-SATB2 |
| hsa-mir-132-prec | 17p13 | −1.67 | 4.17 | NA‡ |
| hsa-mir-150† | 19q13.3 | −1.63 | 5.26 | MYB, SDFR1 |
| hsa-mir-020 | 13q31 | −1.62 | 2.17 | TAL1, SKI, RUNX-1, FLT1, CRK, FOG2, RARB |
| hsa-mir-339 | 7p22 | −1.60 | 3.03 | SKI, ETV6, GATA2, FLT1, RAP1B, JUNB, MEIS2 |
| hsa-let-7a | 9q22 | −1.58 | 2.94 | HOXA1, HOXA9, MEIS2, ITGB3, PLDN |
| hsa-let-7d | 9q22 | −1.56 | 2.17 | HOXA1, HOXD1, ITGB3, RUNX1, PDGFRA |
| hsa-mir-181c | 19p13 | −1.55 | 2.50 | RUNX-1, KIT, HOXA1, MEIS2, ETS-1 ETV6, PDGFRA |
| hsa-mir-181b | 1q31.3 | −1.53 | 2.13 | RUNX-1, KIT, ITGA3, HOXA1, MEIS2, ETS-1, SDFR1. |
| hsa-mir-017 | 13q31 | −1.38 | 1.82 | TAL1, SKI, FLT1, RUNX1, CRK, FOG1, ETS-1, MEIS1 |
All differentially expressed miRNAs have q value <0.01 (false-positive rate).
*t test P < 0.05.
†These miRNAs were identified by PAM as predictors of a megakaryocytic class with the lowest misclassification error. All, except miR-143, are down-regulated during megakaryocytic differentiation.
‡miRNA precursor sequence that not contain the mature miRNA, therefore no putative target is shown.
Fig. 1.
Northern blots and real-time miRNA-PCR validation of miRNA chip data in CD34 progenitor differentiation experiments. (A) Northern blots for miR-223, miR-130a, and miR-10a. Loading RNA control was performed with U6. (B) miRNA RT-PCR for miR-10a, miR-106, miR-126, and miR-130a. The miRNA expression is presented as fold difference with respect to CD34+ cells before culture. (C and D) Temporal array expression of miR-223, miR-15–A, and miR-16-1 by miRNA RT-PCR.
Because we found mainly down-regulation of miRNAs during megakaryocytopoiesis, we hypothesized that these miRNAs may unblock target genes involved in differentiation. In line with this hypothesis, miRNAs that are sharply down-regulated in our system are predicted to target genes with important roles in megakaryocytic differentiation. Among the transcription factors with well known function in megakaryocytopoiesis, RUNX-1 (17), Fli-1 (18), FLT1 (19), ETV6 (20), TAL1 (21), ETS1 (22), and CRK (23) are putative targets for several miRNAs down-regulated in differentiated megakaryocytes. Moreover each of these transcription factors has more than one miRNA predicted to be its regulator. For example, RUNX1 (AML1) is predicted to be the target of miR-106, miR-181b, miR-101, let7d, and the miR-17–92 cluster. The multiplicity of miRNAs predicted to target AML1 suggests a combinatorial model of regulation.
We then looked at the temporal expression of miRNAs during the megakaryocytic differentiation process from CD34+ progenitors. We focused on miRNAs that have been described in hematopoietic tissues, such as miR-223, miR-181, miR-155, miR-142, miR-15a, miR-16, miR-106, and the cluster of miR-17–92 (Fig. 1; see also Fig. 5, which is published as supporting information on the PNAS web site). We found sequential changes in the expression of miR-223: Initially, miR-223 is down-regulated during megakaryocytic differentiation, but after 14 days in culture, its expression returns to levels comparable with that of CD34 progenitors (Fig. 1C). The miR-15a and miR-16-1 cluster also follows the same pattern of expression as miR-223 (Fig. 1D), whereas miR-181b, miR-155, miR-106a, miR-17, and miR-20 were down-regulated during differentiation (Fig. 6, which is published as supporting information on the PNAS web site). The temporal variation of the expression of miR-223 and miR-15a/mir-16-1 suggests a stage-specific function.
MAFB Transcription Factor Is a Target of miR-130a.
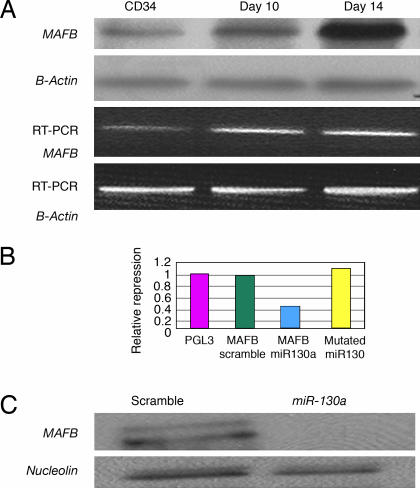
By using three target prediction algorithms [targetscan (http://genes.mit.edu/targetscan), miranda (www.microrna.org/miranda_new.html), and pictar (pictar.bio.nyu.edu)] we identified that miR-130a is predicted to target MAFB, a transcription factor that is up-regulated during megakaryocytic differentiation and induces the GPIIb gene, in synergy with GATA1, SP1, and ETS-1 (24). To investigate this putative interaction, first, we examined MAFB protein and mRNA levels in CD34+ progenitors at baseline and after cytokine stimulation (Fig. 2A). We found that the MAFB protein is up-regulated during in vitro megakaryocytic differentiation. Although the mRNA levels for MAFB by PCR increase with differentiation, this increase does not correlate well with the intensity of its protein expression. The inverse pattern of expression of MAFB and miR-130a suggested in vivo interaction that was further investigated.
Fig. 2.
MAFB is a target of miR-130a. (A) MAFB mRNA and protein expression in CD34+ progenitors induced to megakaryocytic differentiation. β-Actin was used for RT-PCR and Western blot loading controls. (B) Relative repression of luciferase activity in MEG01 cells cotransfected with miR-130A and PGL3 3′UTR MAFB, PGL3 WT, and miR-130 seed match mutated. As a control scramble oligo sequences were cotransfected with PGL3 3′UTR MAFB. (C) Western blotting of MAFB total protein lysates in K562 cells transfected with miR-130a and scramble.
To demonstrate a direct interaction between the 3′ UTRs of MAFB with miR-130a, we inserted the 3′ UTR region predicted to interact with this miRNA into a luciferase vector. This experiment revealed a repression of ≈60% of luciferase activity compared with control vector (Fig. 2B). As an additional control experiment, we used a mutated target mRNA sequence for MAFB lacking five of the complementary bases. As expected, the mutations completely abolished the interaction between miR-130a and its target 3′UTRs (Fig. 2B).
We also determined the in vivo consequences of overexpressing miR-130a on MAFB expression. The pre-miR-130a and a negative control were transfected by electroporation into K562 cells, which naturally express MAFB and lack miR-130a. Transfection of the pre-miR-130a, but not the control, resulted in a decrease in the protein levels at 48 h (Fig. 2C). Northern blotting confirmed successful ectopic expression of miR-130a in K562 cells (Fig. 7, which is published as supporting information on the PNAS web site).
MiR-10a Correlates with HOXB Gene Expression.
It has been reported that in mouse embryos, miR-10a, miR-10b, and miR-196 are expressed in HOX-like patterns (25) and closely follow their “host” HOX cluster during evolution (26). These data suggest common regulatory elements across paralog clusters. MiR-10a is located at chromosome 17q21 within the cluster of the HOXB genes (Fig. 8, which is published as supporting information on the PNAS web site) and miR-10b is located at chromosome 2q31 within the HOXD gene cluster. To determine whether the miR-10a expression pattern correlates with the expression of HOXB genes, we performed RT-PCR for HOXB4 and HOXB5, which are the genes located 5′ and 3′, respectively, to miR-10a in the HOXB cluster. As shown in Fig. 9, which is published as supporting information on the PNAS web site, HOXB4 and HOXB5 expression paralleled that of miR-10a, suggesting a common regulatory mechanism.
MiR-10a Down-Regulates HOXA1.
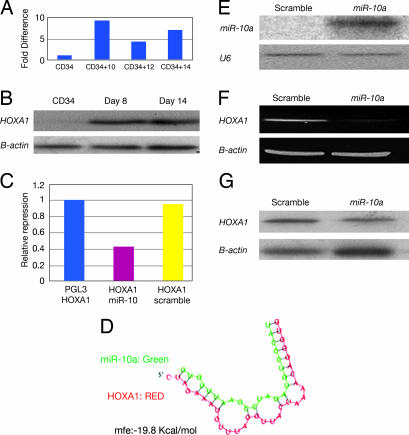
We determined by miRNA array and Northern blot that miR-10a is sharply down-regulated during megakaryocytic differentiation. Interestingly, we found several HOX genes as putative targets for miR-10a (Table 1). We thus investigated whether miR-10a could target a HOX gene. We performed real-time PCR for the predicted HOX targets of miR-10: HOXA1, HOXA3, and HOXD10. After normalization with 18S RNA, we found that HOXA1 mRNA is up-regulated 7-fold during megakaryocytic differentiation compared with CD34 progenitors (Figs. 3A). HOXA1 protein levels were also up-regulated during megakaryocytic differentiation (Fig. 3B). These results are in a sharp contrast with the down-regulation of miR-10a in megakaryocytic differentiation, suggesting that miR-10a could be an inhibitor of HOXA1 expression. To demonstrate a direct interaction of miR-10a and the 3′ UTR sequence of the HOXA1 gene, we carried out a luciferase reporter assay as described in Materials and Methods. When the miRNA precursor miR-10a was introduced in the MEG01 cells along with the reporter plasmid containing the 3′ UTR sequence of HOXA1, a 50% reduction in luciferase activity was observed (Fig. 3C). The degree of complementarity between miR-10a and the HOXA1 3′ UTR is shown in Fig. 3D, as predicted by pictar (http://pictar.bio.nyu.edu).
Fig. 3.
MiR-10a down-regulates HOXA1 by mediating RNA cleavage. (A) RT-PCR for HOXA1 gene expression in differentiated megakaryocytes (relative amount of transcript with respect to CD34+ progenitors at baseline). (B) hoxa1 protein expression in differentiated megakaryocytes. (C) Relative repression of luciferase activity of HOXA1 3′ UTR cloned in PGL3 reporter plasmid when cotransfected with miR-10a and control scramble. (D) Complementarity between miR-10a and the HOXA1 3′UTR as predicted by pictar. (E) Northern blotting for miR-10a gene expression in scramble and miR-10a precursor transfected K562 cells. (F) RT-PCR for HOXA1 gene expression in scramble and miR-10a precursor transfected K562 cells. (G) Western blotting for HOXA1 in K562 cells transfected with control scramble and precursor miR-10a.
To confirm in vivo these findings, we transfected K562 cells with the pre-miR-10a precursor by using nucleoporation and measured HOXA1 mRNA expression by RT-PCR and HOXA1 protein levels by Western blotting. Successful ectopic expression of miR-10a was documented by Northern blot (Fig. 3E). A significant reduction at the mRNA and protein levels for HOXA1 was found for K562 cells transfected with the miR-10a precursor but not with the negative control (Fig. 3 F and G). These data indicate that miR-10a targets HOXA1 in vitro and in vivo.
It has been reported that miR-196 induces cleavage of HOXB8 mRNA, pointing to a posttranscriptional restriction mechanism of HOX gene expression (27). Contrary to the miR-196–HOXB8 interaction, where an almost perfect complementarity exists, the degree of pairing between miR-10a and the human HOXA1 3′ UTR is suboptimal. (Fig. 3D). Although our results indicated target mRNA degradation, further studies are needed to determine whether cleavage or translational repression is the primary mechanism of down-regulation of the HOXA1 gene in this system. A previous study using microarray analysis showed that a large number of target mRNA genes are down-regulated by miRNA at the level of transcription (28). These data raise the question whether target degradation is a consequence of translational repression and subsequent relocalization of the miR–target complexes to cytoplasmic processing bodies or is a primary event (29).
miRNA Profiling in Acute Megakaryoblastic Leukemia (AMKL) Cell Lines.
After the identification of the miRNA expression profile of CD34+ cells during megakaryocytic differentiation, we then investigated miRNA expression in AMKL cell lines with the goal to identify differentially expressed miRNAs that could have a pathogenic role in megakaryoblastic leukemia. We initially compared miRNA expression in four AMKL cell lines with that of in vitro CD34+-differentiated megakaryocytes. Using significance analysis of microarray, we identified 10 miRNAs up-regulated in AMKL cell lines compared with that of CD34 in vitro-differentiated megakaryocytes (Table 2). These miRNAs are as follows (in order of the fold increase with respect to differentiated megakaryocytes): miR-101, miR-126, miR-99a, miR-99-prec, miR-106, miR-339, miR-99b, miR-149, miR-33, and miR-135. Results were validated by RT-PCR as shown in Fig. 10, which is published as supporting information on the PNAS web site. Using PAM, we compared miRNA expression in CD34+ cells with in vitro differentiated megakaryocytes and AMKL cell lines (Table 4, which is published as supporting information on the PNAS web site). Interestingly, we found five miRNAs involved in the megakaryocytic differentiation signature (miR-101, miR-126, miR-106, miR-20, and miR-135) that were up-regulated in the leukemic cell lines (Table 5, which is published as supporting information on the PNAS web site). Whether this profile represents merely a differentiation state of the cells or has a truly pathogenic role remains to be elucidated. Supporting the second hypothesis, miR-106, miR-135, and miR-20 are predicted to target RUNX1, which is one of the genes most commonly associated with leukemia (30). Moreover, mutations of RUNX1 have been described in familial thrombocytopenias with a propensity to develop acute myeloid leukemia (31).
Table 2.
microRNAs up-regulated in acute megakaryoblastic cell lines compared with in vitro-differentiated megakaryocytes
| miRNA | Chromosomal location | t test score | Fold change | Putative targets |
|---|---|---|---|---|
| hsa-mir-101 | 1p31.3 | 6.14 | 11.85 | MEIS2, RUNX1, ETS-1, C-MYB, FOS, RARB, NFE2L2 |
| hsa-mir-126 | 9q34 | 4.91 | 11.97 | V-CRK |
| hsa-mir-099a | 21q21 | 3.30 | 6.83 | HOXA1, EIF2C, FOXA1 |
| hsa-mir-099b-prec | 21q21 | 2.85 | 7.59 | NA |
| hsa-mir-106 | xq26.2 | 2.79 | 3.33 | FLT1, SKI, E2F1, NCOA3, PDGFRA, CRK |
| hsa-mir-339 | 7p22 | 2.58 | 3.36 | HOXA1, FLT1, PTP4A1, RAP1B |
| hsa-mir-099b | 19q13 | 2.46 | 4.19 | HOXA1, MYCBP2 |
| hsa-mir-149 | 2q37 | 2.29 | 3.53 | RAP1A, MAFF, PDGFRA, SP1, NFIB |
| hsa-mir-033 | 2q13 | 2.27 | 3.23 | PDGFRA, HIF1A, MEIS2 |
| hsa-mir-135 | 3p21 | 2.12 | 3.97 | SP1, HIF1A, SP3, HNRPA1, HOXA10, RUNX1 |
All the miRNAs have a q value <0.01 (false-discovery rate). The same miRNAs, except miR-339 and miR-149, were found by using PAM to predict a megakaryoblastic leukemia class with no misclassification error.
Conclusions
In this study, we have found down-regulation of miRNAs during megakaryocytopoiesis. Hypothetically the down-regulation of miRNAs unblocks target genes involved in differentiation. In line with this hypothesis, miRNAs that are sharply down-regulated in our system are predicted to target genes with important roles in megakaryocytic differentiation. Thus, we have shown that miR-130a targets MAFB and miR-10a modulates HOXA1. The fact that we found several differentially expressed miRNAs during differentiation and leukemia that are predicted to target HOXA1 suggests a function for HOXA1 in megakaryocytopoiesis. Loss and gain studies will ultimately be needed to define the role of HOXA1 in this differentiation process. Our findings delineate the expression of miRNAs in megakaryocytic differentiation and suggest a role for miRNA modulation of this lineage by targeting megakaryocytic transcription factors. Furthermore, in megakaryoblastic leukemia cell lines, we have found inverse expression of miRNAs involved in normal megakaryocytic differentiation. These data provide a starting point for future studies of miRNAs in megakaryocytopoiesis and leukemia.
Materials and Methods
Cell Lines and Human CD34+ Cells.
The human chronic myeloid leukemia blast crisis cell lines K-562 and MEG-01 were obtained from American Type Tissue Culture and maintained in RPMI medium 1640 (Gibco) containing 10% FBS with penicillin–gentamycin at 37°C with 5% CO2. The human megakaryoblastic leukemia cells UT-7 and CMK and the chronic myeloid leukemia in blast crisis LAMA were obtained from DSMZ (Braunsweig, Germany). All cells were maintained in RPMI medium 1640 with 20% FBS and antibiotics, except UT-7, which is factor-dependent and was cultured in MEM-α with 20% FBS and 5 ng/ml granulocyte–macrophage colony-stimulating factor. Fresh and frozen human bone marrow CD34+ cells were obtained from Stemcell Technologies (Vancouver, BC, Canada). FACS analysis for CD34 antigen revealed a purity >98%.
Human Progenitor CD34+ Cell Cultures.
Human bone marrow CD34+ cells were grown in STEM media (Stemcell Technologies), which includes Isocove-modified Dulbecco’s medium supplemented with human transferrin, insulin, bovine serine albumin, human low-density lipoprotein, and glutamine, in the presence of 100 ng/ml human recombinant thrombopoietin (TPO) for the first 4 days, followed by a combination of 100 ng/ml TPO, IL3, and stem cell factor (cytokine mixture CC-200, Stemcell Technologies). The initial cell density was 100,000 cells per ml; three times a week, the cell density was adjusted to 100,000 to 200,000 cells per ml. To increase the purity of the cells for microarray analysis, cell sorting was performed at day 10 of culture. Cells were incubated on ice for 45 min with anti-human CD34+, anti-human CD41+, anti-human CD61+, and their respective isotypes. After washing twice with PBS 3% FBS, cells were sorted by using a FACS Aria sorting machine in bulk in two separate populations; CD34− CD61+ and CD34+ CD61+ cells for culture and RNA extraction. The purity of the sorted populations was >95%.
Megakaryocytes Characterization.
Cytospin preparations of CD34+ progenitors in culture were performed and stained with May–Grunwald Giemsa at different time points during the megakaryocytic differentiation induction. For FACS analysis, the primary antibodies used were as follows: CD41A, CD61A, CD42B, and CD34 with their respective isotypes (BD Pharmingen). Cytometric studies were performed as described in ref. 32 by using a FACScalibur (BD Biosciences) and cellquest software (BD Biosciences).
RNA Extraction, Northern Blotting, and miRNA Microarray Experiments.
Procedures were performed as described in detail in ref. 33. Raw data were normalized and analyzed in genespring 7.2 software (zcomSilicon Genetics, Redwood City, CA). Expression data were median-centered by using both the genespring normalization option and the global median normalization of the bioconductor package (www.bioconductor.org) with similar results. Statistical comparisons were done by using the genespring ANOVA tool, predictive analysis of microarray (PAM), and the significance analysis of microarray (sam) software (http://www-stat.stanford.edu/∼tibs/SAM/index.html).
RT-PCR and Real-Time PCR.
Total RNA isolated with TRIzol reagent (Invitrogen) was processed after DNase treatment (Ambion, Austin, TX) directly to cDNA by reverse transcription by using SuperScript II (Invitrogen). Comparative real-time PCR was performed in triplicate. Primers and probes were obtained from Applied Biosystems for the following genes: HOXA1, HOXA3, HOXB4, HOXB5, and HOXD10. Gene expression levels were quantified by using the ABI Prism 7900 Sequence detection system (Applied Biosystems). Normalization was performed by using the 18S RNA primer kit. Relative expression was calculated by using the CT method. RT-PCR also was performed by using the following primers: MAFB FW, 5′-AACTTTGTCTTGGGGGACAC-3′; MAFB RW, 5′-GAGGGGAGGATCTGTTTTCC-′3; HOXA1 FW, 5′-CCAGGAGCTCAGGAAGAAGA GAT-3′; and HOXA1 RW, 5′-CCCTCTGAGGCATCTGATTGGGTTT-3′.
Real-Time Quantification of miRNAs by Stem-Loop RT-PCR.
Real-time PCR for pri-miRNAs 10a, miR15a, miR16-1, miR-130a, miR-20, miR-106, miR-17–5, miR-181b, miR-99a, and miR-126 were performed as described in ref. 34. 18S was used for normalization. All reagents and primers were obtained from Applied Biosystems.
Bioinformatics.
miRNA target prediction of the differentially expressed miRNAs was performed by using targetscan, miranda, and pictar software.
Cell Transfection with miRNA Precursors.
miRNA precursors miR-10a and miR-130a were purchased from Ambion. Five million K562 cells were nucleoporated by using Amaxa (Gaithersburg, MD) with 5 μg of precursor oligos in a total volume of 10 ml. The expression of the oligos was assessed by Northern blots and RT-PCR as described.
Luciferase Reporter Experiments.
The 3′ UTR segments containing the target sites for miR-10a and miR-130a from HOXA1 and MAFB genes, respectively, were amplified by PCR from genomic DNA and inserted into the pGL3 control vector (Promega) by using the XbaI site immediately downstream from the stop codon of luciferase. The following primer sets were used to generate specific fragments: MAFB FW, 5′-GCATCTAGAGCACCCCAGAGGAGTGT-3′; MAFB RW, 5′-GCATCTAGACAAGC ACCATGCGGTTC-3′; HOXA1 FW, 5′-TACTCTAGACCAGGAGCTCAGGAAGA-3′; and HOXA1 RW, 5′-MCATTCTAGATGAGGCATCTGATTGGG-3′. We also generated two inserts with deletions of 5 bp and 9 bp, respectively, from the site of perfect complementarity by using the QuikChange XL-site directed Mutagenesis Kit (Stratagene). WT and mutant inserts were confirmed by sequencing.
Human chronic myeloid leukemia in megakaryoblastic crisis cell line (MEG-01) was cotransfected in six-well plates by using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol with 0.4 μg of the firefly luciferase report vector and 0.08 μg of the control vector containing Renilla luciferase, pRL-TK (Promega). For each well, 10 nM concentrations of the premiR-130a and premiR-10a precursors (Ambion) were used. Firefly and Renilla luciferase activities were measured consecutively by using the dual luciferase assays (Promega) 24 h after transfection.
Western Blots.
Total and nuclear protein extracts from K562 transfected with miR-10a and miR-130a as well as CD34+ cells at different stages of megakaryocytic differentiation were extracted by using RIPA buffer (0.15 mM NaCl/0.05 mM Tris·HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) or a nuclear extraction kit (Pierce). Protein expression was analyzed by Western blotting with the following primary antibodies: MAFB (Santa Cruz Biotechnology), HOXA1 (R & D Systems), β-actin, and Nucleolin (Santa Cruz Biotechnology). Appropriate secondary antibodies were used (Santa Cruz Biotechnology).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Program Project Grants P01CA76259, P01CA16058, and P01CA81534 (to C.M.C.) and P01CA16672 (to M.A.). The Leukemia Clinical Research Foundation supports C.D.B. Both G.A.C. and R.A. are supported by Kimmel Foundation grant awards, and G.A.C. is supported by a Chronic Lymphocytic Leukemia Global Research Foundation grant.
Abbreviations
- AMKL
acute megakaryoblastic leukemia
- miRNA
microRNA
- PAM
predictive analysis of microarray.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Bartel D. P. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Karp X., Ambros V. Science. 2005;310:1330–1333. doi: 10.1126/science.1121566. [DOI] [PubMed] [Google Scholar]
- 4.Cheng A. M., Byrom M. W., Shelton J., Ford L. P. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu P., Guo M., Hay B. A. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Poy M. N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., MacDonald P. E., Pfeffer S., Tuschl T., Rajeswski N., Rorsman P., et al. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 7.Dresios J., Aschrafi A., Owens G. C., Vanderkilsh P. W., Edelman G. M., Mauro V. P. Proc. Natl. Acad. Sci. USA. 2005;102:1865–1870. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin G. A., Dumitru C. D., Shimizu M., Bichi R., Zupo S., Noch E., Adler H., Rattan S., Keating M., Rai K., et al. Proc. Natl. Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin G. A., Liu C.-G., Ferracin M., Sevignani C., Dumitru C. D., Shimizu M., Cimmino A., Zupo C., Rattan S., Dono M., Dell’Aquila M. L., et al. Proc. Natl. Acad. Sci. USA. 2004;101:11755–11760. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He L., Thompsom J. M., Hermann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S., Hanon G. J., Hammond S. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J., Getz G., Miska E. A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B. L., Mak R. H., Ferrando A. A., et al. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 12.Chen C. Z., Li L., Lodish H. F., Bartel D. P. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 13.Monticelli S., Ansel K. M., Xiao C., Socci N., Krichevsky A., Tai T. H., Rajewsky N., Marks D. S., Sander C., Rajewsky K., et al. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Felli N., Fontana L., Pelosi E., Botta R., Bonci D., Fachiano F., Liuzzi F., Lulli V., Morsili O., Santoro S., et al. Proc. Natl. Acad. Sci. USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fazi F., Rosa A., Fatica A., Gelmetti V., Marchis M. L., Nervi C., Bozzoni I. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 16.Metzler M., Wilda M., Busch K., Viehmann S., Borkhardt A. Genes Chromosomes Cancer. 2004;39:167–169. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 17.Elagib K. E., Racke F. K., Mogass M., Khetawat R., Delehanty L. L., Goldfarb A. N. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- 18.Athanasoiu M., Clausen P. A., Mavrothalasitiss G. J., Zhang X. K., Watson D. K., Blair D. G. Cell Growth Differ. 1996;7:1525–1534. [PubMed] [Google Scholar]
- 19.Casella I., Feccia T., Chelucci C., Samoggia P., Castelli G., Guerriero R., Parolini I., Petrucci E., Pelosi E., et al. Blood. 2003;101:1316–1323. doi: 10.1182/blood-2002-07-2184. [DOI] [PubMed] [Google Scholar]
- 20.Hock H., Meade E., Medeiros S., Schindler J. W., Valk P. J., Fujiwara Y., Orkin S. H. Genes Dev. 2004;18:2336–2341. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Begley C. G., Green A. R. Blood. 1999;93:2760–2770. [PubMed] [Google Scholar]
- 22.Jackers P., Szalai G., Moussa O., Watson D. K. J. Biol. Chem. 2004;279:52183–52190. doi: 10.1074/jbc.M407489200. [DOI] [PubMed] [Google Scholar]
- 23.Lannutti B. J., Shim M. H., Blake N., Reems J. A., Drachman J. G. Exp. Hematol. 2003;12:1268–1274. doi: 10.1016/j.exphem.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Sevinsky J. R., Whalen A. M., Ahn N. G. Mol. Cell. Biol. 2004;24:4534–4545. doi: 10.1128/MCB.24.10.4534-4545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mansfield J. H., Harfe B. D., Nissen R., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A. E., Ruvkun G., et al. Nature. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 26.Tanzer A., Amemiya C. T., Kim C. B., Stadler P. F. J. Exp. Zool. B Mol. Dev. Evol. 2005;304:75–85. doi: 10.1002/jez.b.21021. [DOI] [PubMed] [Google Scholar]
- 27.Yekta S., Shih I. H., Bartel D. P. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 28.Lim L. P., Lau N. C., Garret-Engele P., Grimsom A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. Nature. 2005;433:769–771. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 29.Pillai R. RNA. 2005;11:1753–1761. doi: 10.1261/rna.2248605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakao M., Horjike S., Fukushima-Nakase Y., Fujita Y., Masafumi T., Okuda T. Oncogene. 2004;125:709–719. doi: 10.1111/j.1365-2141.2004.04966.x. [DOI] [PubMed] [Google Scholar]
- 31.Song W. J., Sullivan M. G., Legare R. D. Nat. Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 32.Tajima S., Tsuji K., Yoshino H. J. Exp. Med. 1996;184:1357–1364. doi: 10.1084/jem.184.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C.-G., Calin G. A., Meloon B., Gamliel N., Sevignani C., Ferracin M., Dumitru C. D., Shimizu M., Zupo S., Dono M., et al. Proc. Natl. Acad. Sci. USA. 2002;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., et al. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.