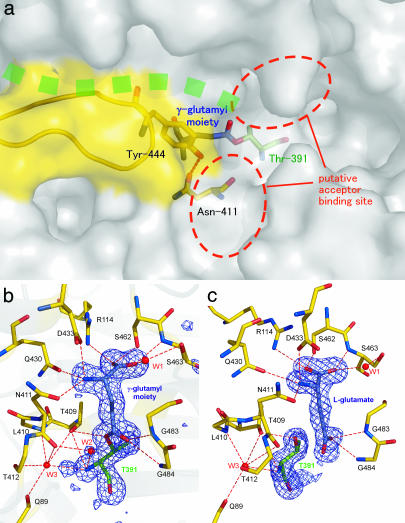
Fig. 2.
The structure of the substrate binding pocket of E. coli GGT. (a) Surface drawing of substrate binding pocket. The stick model of the γ-glutamyl moiety, nucleophile (Thr-391), and residues forming the wall (Asn-411 and Tyr-444) are shown in blue, green, and yellow, respectively. Green dots represent the groove in which the peptide of the precursor protein is assumed to be present. The hydrogen bond between Asn-411 Oδ and Tyr-444 Oη is shown as a dashed line. The ribbon model shown in yellow represents residues Pro-438–Gly-449, which are absent in B. subtilis GGT. (b) The (Fo − Fc) omit map contoured at the 3σ level for GGT-γG. The omit map was generated by omitting the γ-glutamyl moiety, Thr-391, and a water molecule (labeled W2) from the model. Ball-and-stick models of γ-glutamyl–enzyme complex are overlaid on the map. The residues involved in substrate binding and enzyme reaction are shown in the model. For the clarity, the side chains of Gln-89, Leu-410, and Thr-412 are omitted from the model. Water molecules involved in substrate binding and the catalytic reaction are labeled (W1∼W3). The hydrogen bonds are shown as dashed lines. (c) The (Fo − Fc) omit map for GGT-Glu prepared as for GGT-γG. The view direction is rotated by 40° around the vertical axis relative to b. The figures were prepared with pymol (20).