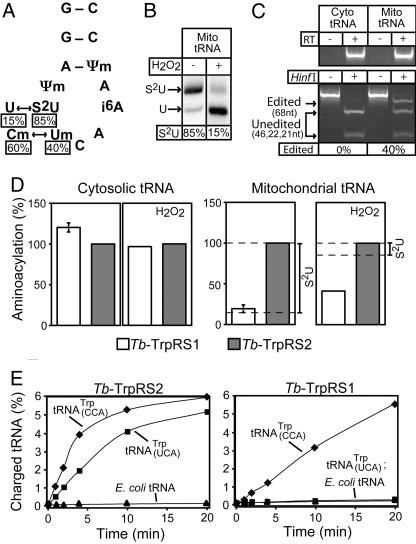
Fig. 5.
Tb-TrpRS1 and Tb-TrpRS2 have distinct substrate specificities. (A) Mitochondrial editing and modification events of the anticodon loop of the tRNATrp as described for Leishmania (14). The percentages of the thiolated U (s2U) and the C→U editing as determined for T. brucei tRNATrp are indicated. (B Left) The percentage of thiomodified mitochondrial tRNATrp of T. brucei was measured by N-acryloylaminophenylmercuric chloride gel electrophoresis (33) and Northern blot hybridization. The shifted band represents thiolated tRNATrp(S2U). (B Right) The fraction of thiolated mitochondrial tRNATrp that remains thiolated after H2O2 treatment was determined as in the left lane. (C) Quantitative RT-PCR assay for edited tRNATrp. (C Upper) The cytosolic and mitochondrial tRNA fractions that were used as templates are free of DNA. (C Lower) The blot shows that RNA editing can be analyzed by a restriction digest because it destroys a HinfI site that is present in the cDNA derived from the unedited tRNATrp (14). Introduction of a synthetic HinfI plus 20 flanking nucleotides at the 5′ end of the 5′ RT-PCR primer provides an internal control for the HinfI digestion, allowing the quantitative determination of RNA editing. cDNA amplified from unedited tRNATrp contains two HinfI sites and, thus, will be digested into three fragments (46, 22, and 21 nt; unedited). The cDNA derived from edited tRNATrp contains the synthetic HinfI site only and will be digested into two fragments (68 and 21 nt; edited). Measuring the intensities of the diagnostic bands for nonedited (46 nt) and edited (68 nt) tRNATrp allows, after correcting for their different molecular mass, determination of the fraction of edited tRNATrp in T. brucei mitochondria. (D) In vitro aminoacylation assays by using [3H]tryptophan and recombinant Tb-TrpRS1 as well as Tb-TrpRS2 as enzymes. (Left and Center Left) Untreated and H2O2-treated cytosolic tRNA were charged. (Right and Center Right) Same as Left and Center Left but with untreated and H2O2-treated mitochondrial tRNAs. For each graph, the tRNA charged by the Tb-TrpRS2 was set to 100%. The percentage of mitochondrial tRNATrp that is thiolated in the untreated and treated fractions is indicated on the right. The means and SE for three independent experiments are shown for aminoacylation of untreated tRNAs. For experiments using H2O2-treated tRNAs, the mean of two experiments is shown. The two values each for cytosolic and mitochondrial tRNAs varied by 20% and 10%, respectively. (E) Aminoacylation of T. brucei tRNATrp overexpressed in E. coli by using 200 nM enzyme and 0.25 μg/μl tRNA. (Right) Tb-TrpRS1 charging of total E. coli tRNA with T. brucei tRNATrpUCA overexpressed (■), total E. coli tRNA with T. brucei tRNATrpCCA overexpressed (♦), and total E. coli tRNA (▴). (Left) Tb-TrpRS2 charging of total E. coli tRNA with T. brucei tRNATrpUCA overexpressed (■), total E. coli tRNA with T. brucei tRNATrpCCA overexpressed (♦), and total E. coli tRNA (▴).