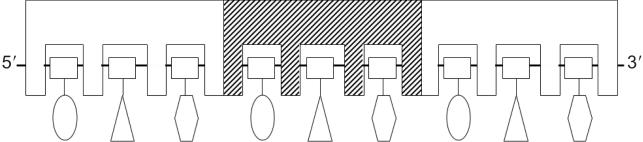
Figure 1.
Model for the binding of RecA to ssDNA. The figure is adapted from Ref. (22). Three RecA monomers are shown, and the ssDNA (9 nt) is bound along the sugar phosphate backbone, which is represented by the solid line and squares. Each RecA monomer has three separate nucleotide-binding sites. Although the bases are primarily exposed for homology recognition (11), in the model they form subtle interactions with the binding sites, giving rise to slight preferences for the different nucleotides and optimal binding to certain trinucleotide-repeating sequences.