Figure 1.
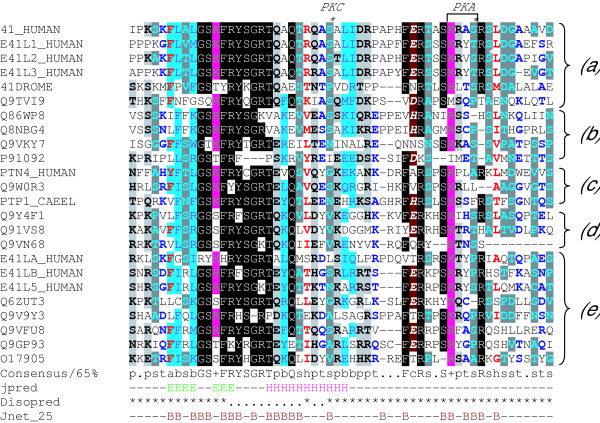
The FERM-adjacent (FA) region. An HMM for the FA region was used to search the Uniprot non-redundant database. The alignment shown here represents sequences detected with the HMM in human, D. melanogaster and C. elegans. The alignment was generated by aligning the sequences to the HMM using HMMALIGN. The sequences shown are all adjacent to the C-terminal end of FERM domains, but not all FERM domains are adjacent to this sequence. Note the presence of a protein kinase A consensus sequence [KR][KR][X][ST] (boxed) in human protein 4.1R (41_HUMAN): the serine (asterisked) in this motif is a physiological PKA substrate. A PKC substrate site is also marked. In 4.1G (E41L2_HUMAN) another serine is phosphorylated in vivo (white asterisk). Secondary structure predictions generated are shown below the alignment. Jpred: E beta-strand; H alpha-helix. Disopred: * disorder. Jnet: B inaccessible to water. The proteins are grouped according to the classes shown in Fig. 2.
