Abstract
The kinetics of forming a proper Watson–Crick base pair as well incorporating bases opposite furan, an abasic site analog, have been well characterized for the B Family replicative DNA polymerase from bacteriophage T4. Structural studies of these reactions, however, have only been performed with the homologous enzyme from bacteriophage RB69. In this work, the homologous enzymes from RB69 and T4 were compared in parallel reactions to determine the relative abilities of the two polymerases to incorporate correct nucleotides as well as to form improper pairings. The kinetic rates for three different exonuclease mutants for each enzyme were measured for incorporation of an A opposite T and an A opposite furan as well as for the formation of A:C and T:T mismatches. The T4 exonuclease mutants were all ∼2- to 7-fold more efficient than the corresponding RB69 exonuclease mutants depending on whether a T or furan was in the templating position and which exonuclease mutant was used. The rates for mismatch formation by T4 were significantly reduced compared with incorporation opposite furan, much more so than the corresponding RB69 mutant. These results show that there are kinetic differences between the two enzymes but they are not large enough to preclude structural assumptions for T4 DNA polymerase based on the known structure of the RB69 DNA polymerase.
INTRODUCTION
The accurate replication of the genome is essential for the survival of all species. The process of accurate DNA replication is carried out by multi-enzyme complexes, the core subunits of which are the replicative DNA polymerases that carry out the actual enzymatic reaction of inserting nucleoside triphosphates opposite template bases. Detailed kinetic mechanisms have been elucidated for several families of DNA polymerases with one of the most intensely studied being that of gene product 43 (gp43) of the bacteriophage T4. Accurate replication is accomplished by selection of the correct nucleotide based on its ability to fit within the active site of the enzyme (1), conformational changes, especially the movement of the fingers domain from an open to a closed position within the protein that form a tight active site (2,3), and an associated 3′–5′ exonuclease activity that allows the enzyme to remove misincorporated nucleotides (4). Together, these activities produce an accuracy rate of one mistake for every 108 nt incorporations for the T4 DNA polymerase, one of the most accurate DNA polymerases known (5,6).
The fidelity of replicative DNA polymerases is put to the test, however, when the enzyme encounters a lesion in the DNA. The most common lesions occurring within DNA are abasic sites, in which the backbone remains intact but the nucleotide base has been lost. These occur at an estimated rate of ∼10 000 per human cell per day (7) mostly by spontaneous depurination [the rates of which are enhanced by bulky adducts such as aflatoxin B1 (8)], by hydroxyl radical attack on the sugar residue (9) and as an intermediate in the base excision repair pathway (10). Abasic sites are strong blocks to DNA replication but can be bypassed by the replicative polymerases with a dependence upon sequence context (11), the presence of processivity factors (12) and the absence of proofreading (13). Because abasic sites are non-coding lesions, often derived from depurination of guanine, successful bypass with purines inserted opposite will result in a ‘non-cognate’ base being incorporated in the majority of instances. Abasic sites can also be bypassed by certain Y family polymerases, but at relatively low efficiencies (14,15). The strong blockage to replication, mutagenic potential, and frequency of formation within the human genome make abasic sites a prime target for the study of the interactions between DNA lesions and DNA polymerases.
The first step in lesion-induced mutagenesis is the initial encounter of a DNA polymerase with the lesion. Kinetics studies have been performed with the replicative DNA polymerase from bacteriophage T4 for both encountering normal bases within the DNA template (16,17) as well as tetrahydrofuran (furan, an abasic site analog), which is the subject of this study (18–20). Structural information for this enzyme, however, is limited to that of the N-terminal domain of the enzyme containing the exonuclease active site (21). A closely related polymerase from bacteriophage RB69 that has 61% sequence identity and 14% sequence similarity to the T4 protein has, however, been crystallized in a number of conformations: the structure of the RB69 gp43 DNA polymerase has been solved as an apo complex (22), an open, editing complex (23), and a closed, ternary complex forming a T:A base pair (3). This enzyme has also been crystallized in the presence of DNA lesions including a ternary complex forming a C:8-oxo-G base pair (24), an open, binary complex with a furan at the templating position (24) and a binary complex in which an A has been incorporated opposite a furan (25).
Using rapid quench methods, we determined comparative kinetic values for insertion opposite a furan for the T4 and RB69 DNA polymerases as an aid in determining the applicability of RB69 structural information to other members of the B Family of DNA polymerases. Both polymerases, in the form of three different exonuclease-deficient mutants, were compared side by side to determine their relative abilities to form both proper A:T pairings and improper A:furan pairings. We show that measurable differences exist between the T4 and RB69 enzymes, especially when a furan is in the templating position. These differences, however, are small and suggest that structural data obtained from the RB69 DNA polymerase are applicable to its homolog from T4.
MATERIALS AND METHODS
Materials
Oligonucleotides were from Midland Certified Reagent Company and consisted of a primer (5′-GAATTCCAGACTGTCACTGG-3′) and template (5′-TTGCGFCCAGTGACAGTCTGGAATTC-3′) strand. The 5′ end of the primer strand was labeled with a TET (Tetrachlorofluorescein) fluorescent marker for subsequent analysis by PAGE. The template contained a tetrahydrofuran residue that is a chemically stable abasic site analog (26) and is the same analog used in previous biochemical (18–20) and structural (24,25) studies. The oligonucleotides were gel purified on 16% polyacrylamide gels, eluted in 500 mM NaCl, de-salted on C18 Sep-Pak cartridges (Waters), and spun to dryness. Resuspended primer and template oligonucleotides were annealed in a buffer containing 10 mM Tris (pH7.5), 50 mM NaCl and 1 mM EDTA by heating to 90°C and cooling to room temperature over several hours. A 20% excess of template was used to ensure maximum annealing of the primer strand.
dATP and dTTP were from Invitrogen and all chemicals were purchased from Sigma.
Protein purification
Exonuclease mutant T4 DNA polymerases (D112A/E114A, D219A and D324A) were constructed and purified as described previously (27).
Dr Jim Karam kindly provided a plasmid containing the wild-type RB69 gp43 DNA polymerase gene with a histidine tag. All three exonuclease-deficient mutants of RB69 (D114A/E116A, D222A and D327A) were generated by creating point mutations following the protocol of the Quickchange XL kit (Stratagene). Wild-type and mutated plasmids were transformed into BL21 (DE3) which were induced with 1 mM isopropyl-β-d-thiogalactopyranoside and grown at 37°C to an A600 of 0.5. Cells were then lysed in a buffer of 50 mM sodium phosphate (pH 8), 100 mM NaCl, 10 mM imidazole (pH 8), 10% glycerol, 5 mM 2-mercaptoethanol (BME), 1 mM phenylmethylsulphonyl fluoride and 10 mM benzamidine. Clarified lysate was then loaded onto a nickel sulfate-charged chelating column (Amersham Biosciences) and eluted with a gradient up to 500 mM imidazole. Pooled fractions were dialyzed into a buffer containing 10% glycerol, 20 mM Tris (pH 7.5), 1 mM EDTA, 10 mM MgCl2, 10 mM BME and 20 mM NaCl and loaded onto a Q column (Amersham Biosciences). Protein was eluted with a gradient to 500 mM NaCl. Pooled fractions were dialyzed into a storage buffer of 50% glycerol, 200 mM potassium phosphate and 10 mM BME and stored at −20°C.
Primer extension reactions
All reactions were performed in an RQF-3 rapid quench apparatus (KinTek). Syringe A contained 2000 nM enzyme and 500 nM DNA substrate in a buffer containing 25 mM Tris–acetate (pH 7.5), 150 mM potassium acetate and 10 mM BME. Syringe B contained 20 mM magnesium acetate and varying concentrations of dATP in an identical buffer. The final concentrations of reactants were 1000 nM enzyme, 250 nM DNA, 10 mM Mg++ and dATP concentrations ranging from 0.002 to 0.4 mM when an A:T base pair was formed and from 0.125 to 4 mM when an A was incorporated opposite furan or an A:C or T:T mismatch was formed.
All reactions were carried out under conditions of enzyme saturation with a 4-fold excess of enzyme over DNA. The DNA was saturated with protein under these conditions since a 6- or 8-fold excess of protein did not lead to faster rates. Reactions involving mispairs (dATP opposite either furan or cytosine and dTTP opposite thymine) were carried out at 25°C while Watson–Crick base pairing (dATP opposite thymine) was measured at 12°C. The reactants were pushed into the reaction chamber using a buffer equal to that in the reagent mixtures and were quenched with 0.5 M EDTA (pH 8).
Data analyses
Ten microliters of each quenched reaction were mixed with ten microliters formamide containing 0.1 % bromophenol blue. Each sample was loaded onto a 16% polyacrylamide gel containing 8 M urea and run at 50 watts in TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA) buffer to separate extension products from unextended primer. Product formation was visualized using a Bio-Rad molecular imager at the Alexis 532 setting to excite the TET fluorophore linked to the 5′ end of the primers. Equal volumes were used to quantify the densities of both the extended and unextended primer bands as well as a background volume for each pair of bands. The background density was subtracted from each band and the fraction extension was calculated by dividing the density of the extended primer band by the sum of both extended and unextended primer bands (28).
Product formation as a function of time was plotted using Prism (GraphPad Software). Curves were best fit using a double exponential equation for incorporation of an A opposite a T, while a single exponential was used to fit the curves of insertion of an A opposite furan or opposite a C and a T opposite a T. Observed rate constants for all experiments were plotted against dATP concentration and fit to a hyperbolic curve (kobs = kpol[dATP]/(KD + [dATP])). Values for kpol and KD values for dATP binding as well as their standard errors were calculated by non-linear regression with the program Prism.
RESULTS
Primer extension assays
We set out to test our hypothesis that RB69 DNA polymerase would exhibit similar kinetics of insertion of an A opposite furan as T4 DNA polymerase. Kinetic values for incorporation of an A opposite furan have previously been determined for T4 DNA polymerase (18), but the crystal structure of an insertion opposite furan has only been determined for its homolog from bacteriophage RB69 (24,25). To demonstrate that the structure of RB69 DNA polymerase inserting an A opposite furan was applicable to other members of the B family of DNA polymerases, we made a direct comparison between the rates for insertion opposite furan for T4 with the unknown rates for RB69. Initial experiments suggested that the version of RB69 we had been using for structural studies (D222A/D327A exonuclease-deficient mutant) showed different kinetic rates for inserting an A opposite furan than the T4 exonuclease mutant we had in the lab (D112A/E114A). Although the differences could have been due to the presence of a histidine tag on the C-terminal tail of the RB69 variants that is not present on the T4 variants, in control experiments the kinetic rates for a non-His-tagged RB69 D327A mutant were indistinguishable from the His-tagged version (data not shown). This, along with unpublished structural studies, suggests that the His-tag is not likely to be influencing the kinetic differences we have observed between the T4 and RB69 DNA polymerases used in this work.
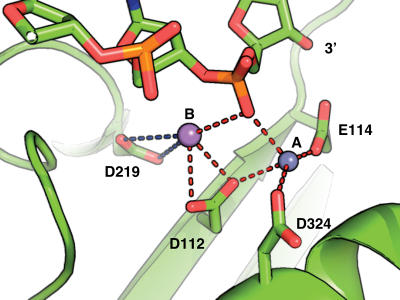
The arrangement of the amino acid residues mutated in the polymerases used in these studies is shown in Figure 1 and is indistinguishable between the T4 and RB69 exonuclease active sites (21,23,25). Different amino acid substitutions in the exonuclease active site have varying effects on polymerase activity (29); hence, we hypothesized that the differences observed for the two enzymes were due to non-equivalent modifications in the exonuclease active site. To test this proposal we measured the kinetic rates of polymerization in parallel for three different exonuclease-deficient mutants (D112A/E114A, D219A, D324A in T4 and the corresponding D114A/E116A, D222A, D327A mutants in RB69).
Figure 1.
Exonuclease active site of T4 DNA polymerase (21) (PDB: 1NOY). The structure of the exonuclease active site is shown with the four conserved catalytic residues. The two divalent metal ions are shown as spheres. Inter-atomic distances <3.5 Å are shown as red, dashed lines and those >3.5 Å are shown in blue. This figure was made with PyMOL (37).
dAMP opposite furan
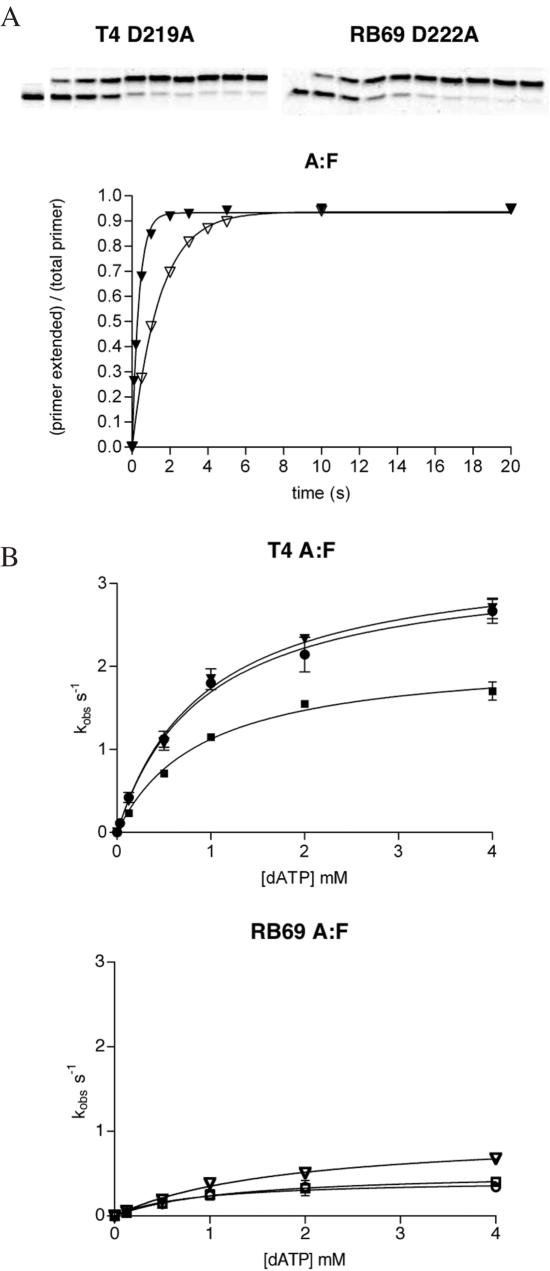
The ability of the various exonuclease mutants to insert a nucleotide opposite furan was tested under enzyme saturating conditions for a single incorporation of an A opposite furan. We chose to use dATP because B Family polymerases follow the ‘A-rule’ in which dAMP is incorporated opposite abasic sites preferentially over the three other canonical nucleotides (30). Enzyme saturating conditions in which the concentration of enzyme is in excess of DNA substrate were used such that the maximum number of primer/templates are bound by a polymerase and can undergo one round of nucleotide incorporation. A 4-fold excess of enzyme satisfied these requirements as adding more enzyme did not yield greater rates of incorporation. The progress curves of primer extension as a function of time were best fit with a single exponential equation when an A was incorporated opposite furan to give values of kobs (Figure 2A). These rate constants were then plotted against nucleotide concentrations and fit to a hyperbola to give values for kpol and the dissociation constant, KD, for dATP (Figure 2B). The results of these experiments show that none of the RB69 exonuclease mutants incorporate dAMP opposite furan as readily as the T4 exonuclease mutants (Table 1). The kpol rates for inserting an A opposite furan are 4-fold greater for T4 D219A versus RB69 D222A as well as for T4 D324A versus RB69 D327A. An 8-fold greater kpol was observed for T4 D112A/E114A when compared with RB69 D114A/E116A. The dissociation constants of dATP binding opposite the furan for all of the mutants, however, were not statistically different from each other. These results suggest that all of the T4 and RB69 variants have an equally low affinity for dATP opposite furan, but the rate limiting step between nucleotide binding and product formation is at least four times faster for T4 than for RB69. Interestingly, the kpol value measured for T4 D219A (3.36 ± 0.1 s−1) and the KD dATP (933 μM) are 22- and 27-fold greater than those measured previously for this T4 mutant forming an A:furan site pairing (0.15 s−1 and 35 μM, respectively) (18). Calculations of the enzyme efficiency (kpol/KD) measured in both experiments, however, differ by <2-fold, well within the range in which differing sequence contexts would be expected to alter the effect of a furan in the template (11).
Figure 2.
Kinetic analysis of dAMP incorporation opposite furan (F). (A) Representative gels and progress curves are shown for the fastest mutants at the highest concentration of dATP used in the experiments (4 mM dATP) plotted against reaction time. These curves were best fit to a single exponential equation []. (B) Calculation of polymerase rate constants and dissociation constants for the incoming nucleotide. The observed rate constants (kobs) for incorporation of A opposite F were plotted against dATP concentrations and fit to a hyperbola to give values for kpol and KD dATP for all three exonuclease mutants of T4 and RB69 DNA polymerases. (closed triangle = T4 D219A, open triangle = RB69 D222A, closed circle = T4 D112A/E114A, open circle = RB69 D114A/E116A, closed square = T4 D324A, open square = RB69 D327A).
Table 1.
Results of primer extension assays
| kpol (s−1) | KD dATP (μM) | kslow(s−1) | |
|---|---|---|---|
| A:T | |||
| T4 D219A pol | 402 ± 14 | 13 ± 2 | 20 ± 2.5 |
| T4 D112A/E114Apol | 361 ± 13 | 11 ± 2 | 19 ± 2.5 |
| T4 D324A pol | 180 ± 17 | 18 ± 9 | 19 ± 1.4 |
| RB69 D222A pol | 320 ± 25 | 42 ± 13 | 13 ± 1.5 |
| RB69 D114A/E116A pol | 270 ± 27 | 46 ± 16 | 13 ± 1.2 |
| RB69 D327A pol | 224 ± 20 | 47 ± 20 | 14 ± 1.9 |
| A:F | |||
| T4 D219A pol | 3.36 ± 0.18 | 934 ± 152 | |
| T4 D112A/E114A pol | 3.24 ± 0.23 | 915 ± 210 | |
| T4 D324A pol | 2.13 ± 0.11 | 897 ± 146 | |
| RB69 D222A pol | 0.97 ± 0.07 | 1767 ± 314 | |
| RB69 Dl14A/E116A pol | 0.43 ± 0.04 | 823 ± 280 | |
| RB69 D327A pol | 0.53 ± 0.09 | 1210 ± 523 | |
| A:C | kobs (s−1) | ||
| T4 D219A pol | 0.56 | ||
| RB69 D222A pol | 0.43 | ||
| T:T | kobs (s−1) | ||
| T4 D219A pol | 0.18 | ||
| RB69 D222A pol | 0.04 | ||
| kpol/Kd (s−1μM−1) | A:T | A:F | |
| T4 D219A pol | 30.9 ± 4.9 | 3.59 ± 0.31 × 10−3 | |
| T4 D112A/E114A pol | 32.8 ± 6.1 | 3.50 ± 0.87 × 10−3 | |
| T4 D324A pol | 10.0 ± 5.1 | 2.37 ± 0.42 × 10−3 | |
| RB69 D222A pol | 7.6 ± 2.4 | 0.55 ± 0.11 × 10−3 | |
| RB69 D114A/E116A pol | 5.9 ± 2.1 | 0.52 ± 0.18 × 10−3 | |
| RB69 D327A pol | 4.8 ± 2.1 | 0.44 ± 0.20 × 10−3 | |
For Watson–Crick base pairs (A:T), values for the fast rate constant, kpol, the dissociation constant of dATP, KD, and the slow rate constant, kslow, are listed. For misincorporations of an A opposite a furan (A:F), the maximal rate constant, kpol, and dissociation constant for dATP, KD, are listed. The mismatches (A:C and T:T) only show the kobs value for the highest concentration of nucleotide (4 mM). All values are the result of non-linear regression analyses of at least three independent measurements except for the kobs values for the mismatches, which are shown as the average of two experiments. The efficiency of A:T and A:F reactions were calculated by dividing kpol by KD for each polymerase. All ± values are SDs.
dAMP opposite thymine
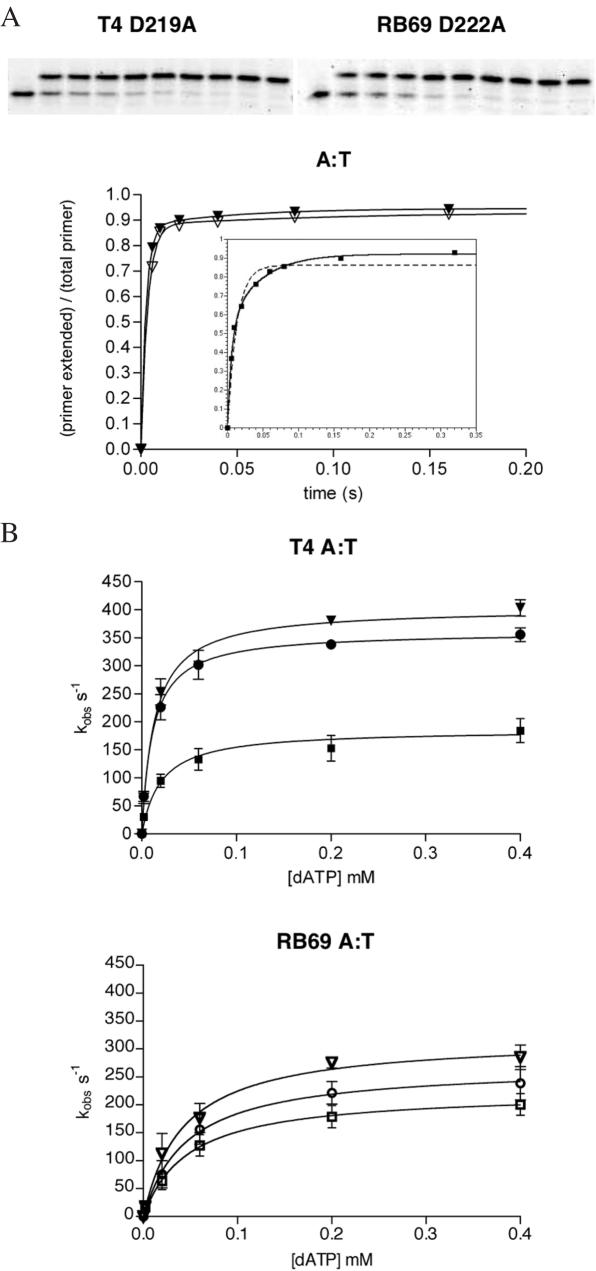
A similar set of experiments were performed using a templating T in place of the abasic site to determine if the observed kpol differences for incorporation opposite furan were due to the presence of the lesion or to intrinsic differences between T4 and RB69 DNA polymerases. All of the progress curves for insertion of an A opposite the templating T were best fit to a double exponential equation that gave both a fast and slow rate constant (Figure 3A and Table 1). The fast rate constants were plotted against dATP concentrations and fit to a hyperbolic function to give values for kpol and dissociation constant, KD, for dATP (Figure 3B). The measured values in this experiment fall into the range of values (100–400 s−1 for kpol and 10–69 μM for KD dNTP) observed previously for normal Watson–Crick base pairs formed with different T4 or RB69 exonuclease-deficient polymerases under varying experimental conditions (1,16,31–33). In contrast to incorporation of an A opposite furan, the rates for an A:T base pair formation are much more similar between the RB69 and T4 variants. The polymerization rates for the RB69 D114A/E116A and the D222A mutants are within 75% of their counterparts in T4 (D112A/E114A and D219A), while the rates for the two slowest mutants, RB69 D327A and T4 D324A, were the same within experimental error. The dissociation constants for dATP were slightly higher for the RB69 mutants but did not differ with statistical significance from those measured for the equivalent T4 mutants.
Figure 3.
Kinetic analysis of dAMP incorporation opposite a templating T. (A) Representative gels and progress curves are shown for the fastest mutants at the highest concentration of dATP used in the experiments (0.4 mM dATP) plotted against reaction time. These curves were best fit to a double exponential equation []. The inset shows the progress curve for the slowest T4 mutant (D324A) and a comparison between a double exponential curve fit (solid line) and a single exponential curve fit (dashed line). (B) Calculation of polymerase rate constants and dissociation constants for the incoming nucleotide. The observed, fast rate constants (kobs) for incorporation of A opposite T were plotted against dATP concentrations and fit to a hyperbola to give values for kpol and KD dATP for all three exonuclease mutants of T4 and RB69 DNA polymerases. (Closed triangle = T4 D219A, open triangle = RB69 D222A, closed circle = T4 D112A/E114A, open circle = RB69 D114A/E116A, closed square = T4 D324A, open square = RB69 D327A.).
When a normal Watson–Crick base pair is formed, the largest difference between the two enzymes was the dissociation constant (KD) of dATP, with all the T4 mutants binding the incoming nucleotide about two to four times more tightly than the corresponding exonuclease mutants in RB69. When the templating base is missing, however, the most significant difference among the proteins was in the values for kpol. To account for the differences in these two variables, rate and dissociation constants, the efficiency of each exonuclease mutant was calculated as the ratio of kpol over KD. These values clearly show that the T4 exonuclease mutants performed better than the RB69 exonuclease mutants by a factor of 2–5 when an A is added opposite a T and by a factor of 6–7 when an A is added opposite furan (Table 1). The results also show that the D324A mutant in T4 is less efficient for both A:T and A:furan site formation, while the D219A and D112A/E114A mutations in T4 showed almost identical efficiencies for both reactions. The equivalent D327A mutant in RB69 had a small, but reproducible, reduction in efficiency for A:T formation when compared with the other two RB69 exonuclease mutants (D114A/E116A, D222A), but was equally as efficient when furan was in the templating position.
Mismatches
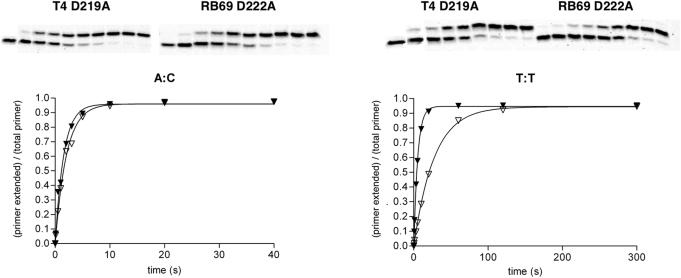
A final experiment was performed to see if the differences observed between the T4 and RB69 mutants when adding an A opposite furan were specific to this particular lesion or were a general response to DNA misincorporations. In this experiment, the templating position for primer extension was either a C or a T and the incoming nucleotide was either an A or a T, respectively, and the polymerases were allowed to form an A:C or T:T mismatch. Only the two fastest mutants (D219A in T4 and D222A in RB69) and only the highest concentration (4 mM) of dATP or dTTP were used (Figure 4). Both mutants incorporated dAMP opposite template C at about the same rate (0.43 and 0.56 s−1), but dTMP was incorporated opposite template T with more difficulty: 0.18 s−1 for the T4 D219A-DNA pol and 0.04 s−1 for the RB69 D222A-DNA pol (Table 1). In this experiment the observed rate constant for RB69 was reduced only slightly from that with an A opposite furan (the kobs value for RB69 D222A at 4 mM dATP was 0.43 s−1 for incorporation of A opposite C and 0.68 s−1 for incorporation opposite furan), while the observed rate constant for T4 D219A was reduced almost 5-fold from that observed when furan was in the templating position (the kobs value for T4 D219A at 4 mM dATP was 0.56 s−1 for incorporation of A opposite C but 3.0 s−1 for incorporation of A opposite furan) and is now quite similar to that of the equivalent RB69 mutant (Table 1).
Figure 4.
Kinetic analysis of mismatch formation. The progress curves for incorporating an A opposite C or T opposite T are shown as the percentage of primer extension plotted against reaction times. Both curves were fit to a single exponential equation []. (Closed triangle = T4 D219A, open triangle = RB69 D222A.).
DISCUSSION
In this study, we compared the ability of exonuclease-deficient RB69 and T4 DNA polymerases to incorporate dAMP opposite furan, a chemically stable abasic site analog, and to form A:C and T:T mismatches. While we originally speculated that differences in the abilities of exo-deficient RB69 and T4 DNA polymerases to incorporate dAMP opposite furan were due to non-equivalent modifications in the exonuclease active center, this proposal was not confirmed by studies reported here. Instead, we find that there are intrinsic differences between the two phage DNA polymerases in their ability to replicate both undamaged and damaged DNA. The mutant T4 DNA polymerases were 6- to 7-fold more efficient in incorporating dAMP opposite furan than the mutant RB69 DNA polymerases (Table 1). The T4 D219A-DNA pol also incorporated dTMP opposite template T ∼4-fold faster than the corresponding RB69 D222A-DNA pol, but both mutant enzymes formed A:C mismatches at about the same rate (Table 1). Interestingly, the T4 D219A-DNA pol incorporated dAMP opposite template C in the presence of 4 mM dATP more slowly (kobs = 0.56 s−1; Table 1) than the incorporation of dAMP opposite furan (kobs = 2.5 s−1; Figure 3B) under the same conditions. In contrast, the corresponding RB69 D222A-DNA pol formed A:C mismatches and incorporated dAMP opposite furan at similar rates (0.43 and 0.5 s−1, respectively; Table 1 and Figure 3B). Another difference observed is that the T4 D219A- and D112A/E114A-DNA polymerases appeared to be more proficient than the corresponding mutant RB69 DNA polymerases in incorporating correct nucleotides (dAMP opposite template T).
The high level of sequence identity between T4 and RB69 DNA polymerases and the fact that the RB69 polymerase can perform accurate replication of the T4 genome during Escherichia coli infection (34) lead to the hypothesis that these proteins should exhibit the same enzymatic behavior in vitro. Mutation rates observed in vivo and in vitro, though, showed small but reproducible differences between DNA polymerases from T4 and RB69 (34). It is not surprising, therefore, that our kinetic data show that there are significant differences between the two proteins in terms of nucleotide incorporation, especially when incorporating opposite furan. A structural basis for these observed differences, however, remains elusive. Threading of the amino acid sequence of T4 DNA polymerase onto the structure of RB69 suggests that all the residues within 5 Å of the incoming nucleoside triphosphate are identical between the two proteins and most of the direct protein interactions with the duplex DNA are by identical residues. Of the residues that differ between the two enzymes, most of these are either in the ‘guide rail’ along the thumb domain that associates with the duplex DNA and are far from the polymerase active site (RB69 residues T703, S783, S784 and P799) or are only remotely associated with the DNA backbone via their main chain atoms (RB69 residues Q356, P390). Thus few structural clues are available for interpreting the differences in binding constants for the incoming nucleoside triphosphate.
A plausible explanation for the reduced polymerization rates for the T4 D324A and RB69 D327A mutants when incorporating an A opposite T is that these mutants favor the formation of complexes with DNA in the exonuclease site. Previous fluorescence studies demonstrate that T4 D219A does not readily form exonuclease complexes (35), but T4 D324A forms exonuclease complexes as well as as observed for the wild-type enzyme (L. Reha-Krantz, unpublished data). Thus differences in the formation of exonuclease complexes could explain the differing kinetic rates observed for the polymerase mutants as well as represent the source of the slow rate constant observed during rapid formation of A:T base pairs. While it is tempting to believe that partitioning and switching of the primer DNA between exonuclease and polymerase active sites can account for the kinetic variation observed in this work, we cannot rule out other polymerase/DNA interactions that can result in a non-extendable conformation (36). The characterization of this interaction will be the focus of future experiments.
The formation of mismatches also suggests a path for future experiments. The current results suggest that the two proteins have subtle structural differences that influence how they see mistakes within the DNA. Previous work has shown that in running start reactions, wild-type T4 DNA polymerase forms different mismatches with varying efficiencies (17). We reiterate this point in standing start reactions with exonuclease-deficient T4 DNA polymerases in that our measured kobs for forming an A:C mismatch is ∼6-fold lower, while forming a T:T mismatch is ∼15-fold lower than that of forming an A:furan. Also of interest is the relative affect on the two different polymerases. The ratio of kobs for A:C mismatch formation by the T4 D219A versus RB69 D222A is almost identical to the kobs ratio for normal A:T base pair formation (1.2 for A:C and 1.4 for A:T), while the kobs ratio for T:T mismatch formation by the two enzymes is similar to the formation of an A:furan (4.5 for T:T and 4 for A:furan). Thus there appears to be an observable difference between the T4 and RB69 DNA polymerases in terms of how readily they make different mistakes. Since there is such conservation of residues between the two polymerases within the immediate environs of the DNA in the polymerase active site, we predict that the observed differences in rate constants for the two proteins will be due to subtle conformational differences between the two proteins. One good candidate would be the volume of the polymerase active sites. Polymerases that are specific for translesion synthesis past bulky lesions as well as small lesions such as abasic sites tend to have larger and more solvent exposed active sites than their replicative counterparts (14). We suspect, then, that subtle differences in active site volumes between the two polymerases may be the determining factor in how well the two polymerases see a variety of DNA mistakes.
This work shows that, despite a high level of sequence conservation, T4 and RB69 replicative DNA polymerases exhibit differing abilities to form various base pairs. Formation of Watson–Crick base pairs occurs at similar rates between the two proteins but the incoming nucleotides are bound less tightly by RB69 DNA polymerase. Incorporation of an A opposite furan by T4 DNA polymerase is more rapid than for RB69 with the two proteins having similar binding constants for the incoming dATP. An A:C mismatch is formed almost equally well by both proteins, while a significant difference exists when a T:T mismatch is formed. While these differences are significant, we feel that they are caused by only subtle rearrangements within the active sites of the two proteins and that structural information garnered from studies with RB69 DNA polymerase is applicable to the T4 DNA polymerase.
Acknowledgments
The authors would like to thank Drs Jeffery Bond, Chris Francklyn and Scott Morrical for technical advice and Dr Stephen Everse for critical reading of this manuscript. This work was supported by NIH R01 CA52040 awarded by the National Cancer Institute to S.S.W. and by grant support to L.R.-K. from the Natural Sciences and Engineering Research Council of Canada. Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1.Capson T.L., Peliska J.A., Kaboord B.F., Frey M.W., Lively C., Dahlberg M., Benkovic S.J. Kinetic characterization of the polymerase and exonuclease activities of the gene 43 protein of bacteriophage T4. Biochemistry. 1992;31:10984–10994. doi: 10.1021/bi00160a007. [DOI] [PubMed] [Google Scholar]
- 2.Doublié S., Tabor S., Long A.M., Richardson C.C., Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 3.Franklin M.C., Wang J., Steitz T.A. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 4.Muzyczka N., Poland R.L., Bessman M.J. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J. Biol. Chem. 1972;247:7116–7122. [PubMed] [Google Scholar]
- 5.Drake J.W. Comparative rates of spontaneous mutation. Nature. 1969;221:1132. doi: 10.1038/2211132a0. [DOI] [PubMed] [Google Scholar]
- 6.Kunkel T.A., Loeb L.A., Goodman M.F. On the fidelity of DNA replication. The accuracy of T4 DNA polymerases in copying phi X174 DNA in vitro. J. Biol. Chem. 1984;259:1539–1545. [PubMed] [Google Scholar]
- 7.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 8.Foster P.L., Eisenstadt E., Miller J.H. Base substitution mutations induced by metabolically activated aflatoxin B1. Proc. Natl Acad. Sci. USA. 1983;80:2695–2698. doi: 10.1073/pnas.80.9.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breen A.P., Murphy J.A. Reactions of oxyl radicals with DNA. Free Radic. Biol. Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 10.Wallace S.S. In: Oxidative Stress and The Molecular Biology of Antioxidant Defenses. Scandalios J., editor. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 49–90. [Google Scholar]
- 11.Hatahet Z., Zhou M., Reha-Krantz L.J., Ide H., Morrical S.W., Wallace S.S. In vitro selection of sequence contexts which enhance bypass of abasic sites and tetrahydrofuran by T4 DNA polymerase holoenzyme. J. Mol. Biol. 1999;286:1045–1057. doi: 10.1006/jmbi.1998.2520. [DOI] [PubMed] [Google Scholar]
- 12.Shibutani S., Takeshita M., Grollman A. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the ‘A rule’. J. Biol. Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 13.Tanguy Le Gac N., Delagoutte E., Germain M., Villani G. Inactivation of the 3′–5′ exonuclease of the replicative T4 DNA polymerase allows translesion DNA synthesis at an abasic site. J. Mol. Biol. 2004;336:1023–1034. doi: 10.1016/j.jmb.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Hogg M., Wallace S.S., Doublie S. Bumps in the road: how replicative DNA polymerases see DNA damage. Curr. Opin. Struct. Biol. 2005;15:86–93. doi: 10.1016/j.sbi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Boudsocq F., Iwai S., Hanaoka F., Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic poleta. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frey M.W., Nossal N.G., Capson T.L., Benkovic S.J. Construction and characterization of a bacteriophage T4 DNA polymerase deficient in 3′→5′ exonuclease activity. Proc. Natl Acad. Sci. USA. 1993;90:2579–2583. doi: 10.1073/pnas.90.7.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Creighton S., Goodman M.F. Gel kinetic analysis of DNA polymerase fidelity in the presence of proofreading using bacteriophage T4 DNA polymerase. J. Biol. Chem. 1995;270:4759–4774. doi: 10.1074/jbc.270.9.4759. [DOI] [PubMed] [Google Scholar]
- 18.Berdis A.J. Dynamics of translesion DNA synthesis catalyzed by the bacteriophage T4 exonuclease-deficient DNA polymerase. Biochemistry. 2001;40:7180–7191. doi: 10.1021/bi0101594. [DOI] [PubMed] [Google Scholar]
- 19.Hays H., Berdis A.J. Manganese substantially alters the dynamics of translesion DNA synthesis. Biochemistry. 2002;41:4771–4778. doi: 10.1021/bi0120648. [DOI] [PubMed] [Google Scholar]
- 20.Reineks E.Z., Berdis A.J. Evaluating the contribution of base stacking during translesion DNA replication. Biochemistry. 2004;43:393–404. doi: 10.1021/bi034948s. [DOI] [PubMed] [Google Scholar]
- 21.Wang J., Yu P., Lin T.C., Konigsberg W.H., Steitz T.A. Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions. Biochemistry. 1996;35:8110–8119. doi: 10.1021/bi960178r. [DOI] [PubMed] [Google Scholar]
- 22.Wang J., Sattar A.K., Wang C.C., Karam J.D., Konigsberg W.H., Steitz T.A. Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 23.Shamoo Y., Steitz T.A. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 24.Freisinger E., Grollman A.P., Miller H., Kisker C. Lesion (in)tolerance reveals insights into DNA replication fidelity. EMBO J. 2004;23:1494–1505. doi: 10.1038/sj.emboj.7600158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogg M., Wallace S.S., Doublié S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeshita M., Chang C.N., Johnson F., Will S., Grollman A.P. Oligodeoxynucleotides containing synthetic abasic sites. Model substrates for DNA polymerases and apurinic/apyrimidinic endonucleases. J. Biol. Chem. 1987;262:10171–10179. [PubMed] [Google Scholar]
- 27.Reha-Krantz L.J., Nonay R.L., Stocki S. Bacteriophage T4 DNA polymerase mutations that confer sensitivity to the PPi analog phosphonoacetic acid. J. Virol. 1993;67:60–66. doi: 10.1128/jvi.67.1.60-66.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boosalis M.S., Petruska J., Goodman M.F. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]
- 29.Reha-Krantz L.J., Nonay R.L. Genetic and biochemical studies of bacteriophage T4 DNA polymerase 3′→5′-exonuclease activity. J. Biol. Chem. 1993;268:27100–27108. [PubMed] [Google Scholar]
- 30.Randall S.K., Eritja R., Kaplan B.E., Petruska J., Goodman M.F. Nucleotide insertion kinetics opposite abasic lesions in DNA. J. Biol. Chem. 1987;262:6864–6870. [PubMed] [Google Scholar]
- 31.Mandal S.S., Fidalgo da Silva E., Reha-Krantz L.J. Using 2-aminopurine fluorescence to detect base unstacking in the template strand during nucleotide incorporation by the bacteriophage T4 DNA polymerase. Biochemistry. 2002;41:4399–4406. doi: 10.1021/bi015723p. [DOI] [PubMed] [Google Scholar]
- 32.Zakharova E., Wang J., Konigsberg W. The activity of selected RB69 DNA polymerase mutants can be restored by manganese ions: the existence of alternative metal ion ligands used during the polymerization cycle. Biochemistry. 2004;43:6587–6595. doi: 10.1021/bi049615p. [DOI] [PubMed] [Google Scholar]
- 33.Yang G., Franklin M., Li J., Lin T.C., Konigsberg W. Correlation of the kinetics of finger domain mutants in RB69 DNA polymerase with its structure. Biochemistry. 2002;41:2526–2534. doi: 10.1021/bi0119924. [DOI] [PubMed] [Google Scholar]
- 34.Dressman H.K., Wang C.C., Karam J.D., Drake J.W. Retention of replication fidelity by a DNA polymerase functioning in a distantly related environment. Proc. Natl Acad. Sci. USA. 1997;94:8042–8046. doi: 10.1073/pnas.94.15.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloom L.B., Otto M.R., Eritja R., Reha-Krantz L.J., Goodman M.F., Beechem J.M. Pre-steady-state kinetic analysis of sequence-dependent nucleotide excision by the 3′-exonuclease activity of bacteriophage T4 DNA polymerase. Biochemistry. 1994;33:7576–7586. doi: 10.1021/bi00190a010. [DOI] [PubMed] [Google Scholar]
- 36.Wohrl B.M., Krebs R., Goody R.S., Restle T. Refined model for primer/template binding by HIV-1 reverse transcriptase: pre-steady-state kinetic analyses of primer/template binding and nucleotide incorporation events distinguish between different binding modes depending on the nature of the nucleic acid substrate. J. Mol. Biol. 1999;292:333–344. doi: 10.1006/jmbi.1999.3057. [DOI] [PubMed] [Google Scholar]
- 37.DeLano W.L. The PyMOL Molecular Graphics System. CA, USA: DeLano Scientific, San Carlos; 2002. [Google Scholar]