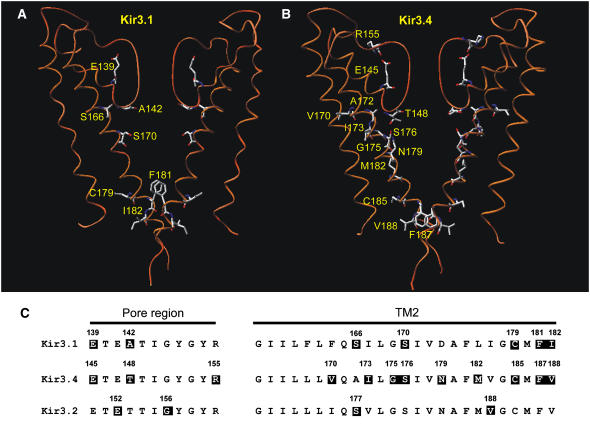
FIGURE 13.
Summary of residues involved in agonist activation of Kir3 channels. (A and B) Models of the TM1-P-TM2 domains of the two Kir3.1 (A) or two Kir3.4 (B) subunits of the Kir3.1/Kir3.4 tetramer based on the KcsA crystal structure. Residues that when mutated abolish agonist activation are highlighted. (C) An alignment of the pore region (left) and TM2 domain (right) in Kir3.1, Kir3.4, and Kir3.2. Residues that when mutated abolish agonist activation are also highlighted. Kir3.1-E-139 (this study); Kir3.1-A-142 (this study); Kir3.1-S-166 (this study); Kir3.1-S-170 (14,16); Kir3.1-C-179 (14); Kir3.1-F-181 (16); Kir3.1-I-182 (14); Kir3.4-E-145 (16); Kir3.4-T-148 (this study); Kir3.4-R-155 (16); Kir3.4-V-170 (13); Kir3.4-I-173 (13); Kir3.4-G-175 (13); Kir3.4-S-176 (13,16); Kir3.4-N-179 (13); Kir3.4-M-182 (13); Kir3.4-C-185 (14); Kir3.4-F-187 (16); Kir3.4-V-188 (14); Kir3.2-E-152 (15); Kir3.2-G-156 (15); Kir3.2-S-177 (15); and Kir3.2-V-188 (15).