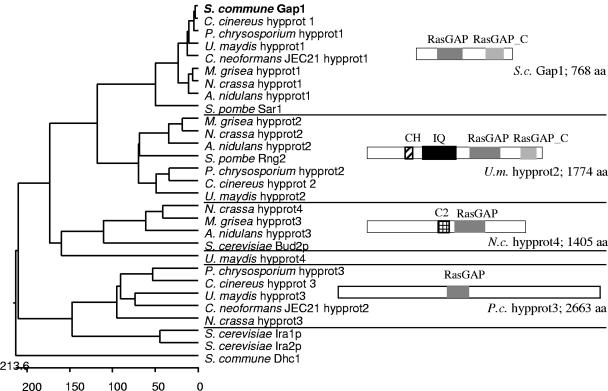
FIG. 2.
Phylogenetic tree of known and putative Ras specific GAPs that were found by BLAST analysis against eight fungal genomes using the S. commune Gap1 sequence. Protein IDs: Aspergillus nidulans hypprot1, EAA61076.1; A.n. hypprot2, EAA66795.1; A.n. hypprot3, EAA59943.1; Cryptococcus neoformans JEC21 hypprot1, EAL17992.1; C.n. hypprot2, CNF01820; Magnaporthe grisea hypprot1, EAA50087.1; M.g. hypprot2, EAA52105.1; M.g. hypprot3, EAA57136.1; Neurospora crassa hypprot1, EAA30748.1; N.c. hypprot2, EAA35739.1; N.c. hypprot3, EAA27028.1; N.c. hypprot4, EAA26628.1; Schizosaccharomyces pombe Sar1, NP_595370.1; S.p. Rng2, O14188; Saccharomyces cerevisiae Bud2p, NP_612831.1; S.c. Ira1p, NP_009698.1; S.c. Ira2p, NP_014560.1; Ustilago maydis hypprot1, EAK81710.1; U.m. hypprot2, EAK86071.1; U.m. hypprot3, EAK82145.1; and U.m. hypprot4, EAK85899.1. Coprinus cinereus and Phanerochaete chrysosporium ORFs were found in the following: C.c. hypprot1, cont1_192 scf9; C.c. hypprot2, cont1_198 scf10; C.c. hypprot3, cont1_258 scf15; P.c. hypprot1, scf31; P.c. hypprot2, scf20; and P.c. hypprot3, scf43. CH, calponin homology domain; IQ, IQ calmodulin-binding motif.