Figure 3.
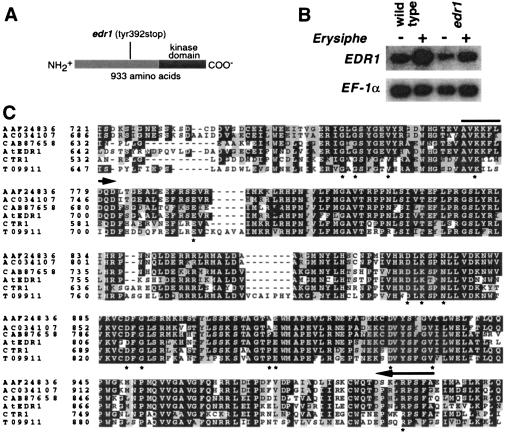
EDR1 encodes a MAPKKK with high similarity to CTR1 and four additional kinases present in Arabidopsis. (A) The predicted EDR1 protein contains 933 amino acids, the carboxyl-terminal third of which contains a kinase domain. (B) RNA gel blot analysis of the EDR1 mRNA. Wild-type and edr1 mutant plants were inoculated with E. cichoracearum (+) or mock inoculated (−) and poly(A) RNA isolated from leaves 3 days after inoculation. The blot was hybridized with an EDR1 cDNA probe corresponding to amino acid residues 180–461. Relative amounts of RNA loaded in each lane were estimated by using an EF-1α cDNA probe. PhosphorImager quantitation of this blot, normalized to the EF-1α signal, revealed that the edr1 message is induced approximately 5-fold by infection with E. cichoracearum. This analysis was repeated three times with similar results. (C) Alignment of the kinase domains of the six known members of the CTR1 kinase family in Arabidopsis. The entire protein for each of the indicated proteins was aligned by using the default parameters of clustalx, a Macintosh version of the clustalw program (47). Only the kinase domain is shown. The full-length alignment can be viewed in supplemental Fig. 6. The alignment file produced by clustalx was formatted by using the boxshade www server (http://www.ch.embnet.org/software/BOX_form.html). White type on a black background indicates a residue that is identical in at least half of the proteins shown, whereas a gray background indicates conservative substitutions as defined by the boxshade 3.21 default parameters. Asterisks indicate residues that are conserved in nearly all protein kinases (48). Arrows indicate regions conserved in the Arabidopsis, tomato, and rice EDR1 orthologs (see supplemental Fig. 6) but divergent in paralogs. Accession nos. for CTR1 and EDR1 are A45178 and AF305913, respectively. Accession no. AC034107 refers to the BAC DNA sequence, from which we derived the indicated protein sequence by using the EDR1 and CTR1 sequences as guides.