FIG.2.
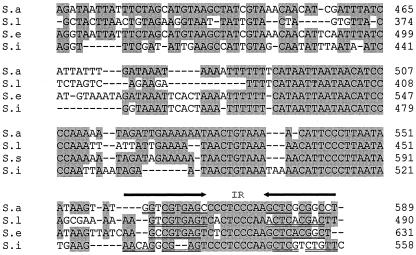
Nucleotide sequence alignment of the RNAIII of S. aureus (S.a), S. lugdunensis (S.l), S. epidermidis (S.e), and S. intermedius (S.i). Identical nucleotides that are present in at least three of the sequences are indicated by shading. The predicted delta-lysin open reading frames (ATG) and the stop codons (TAA) are in boldface type. The putative ribosome-binding Shine-Dalgarno (SD) sites upstream of the delta-lysin are underlined. Arrowheads and double-underlined sequences indicate direct (DR) and indirect repeat (IR). Sequences are manipulated by introducing gaps (indicated by dashes) to fit transcription start points and the delta-lysin open reading frames. Putative transcriptional start sites (+1) and the −10 and −35 promoter elements are underlined.