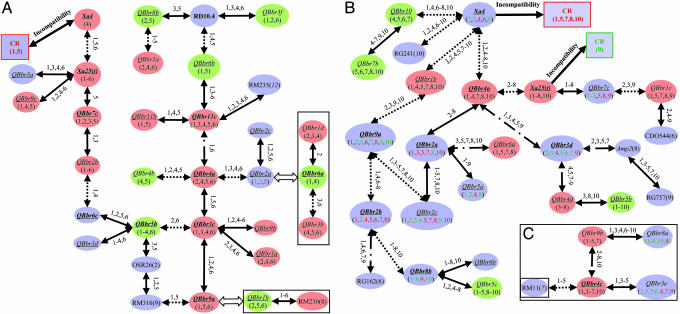
Fig. 2.
Putative genetic networks underlying the defensive system of rice based on the epistatic relationships between R genes and QRL detected. (A) The putative genetic networks underlying CR and PR to six Philippine Xoo races detected in the Lemont/Teqing RILs. (B) The putative genetic networks underlying CR and PR to 10 Philippine Xoo races detected in the IR64/Azucena DHLs. QRL in orange and green represent loci at which the indica (Teqing or IR64) and japonica (Lemont or Azucena) alleles, respectively, resulted in resistance, whereas those in light blue present cases for which resistance resulted from alleles of either parent, depending on Xoo races. Underlined QRL are those detected in both populations and are inferred based on the common markers using the comparative mapping approach (9). The numbers under each QRL represent the Xoo races against which the QRL was effective (Fig. 1). Lines connecting two QRL indicate that significant epistasis was detected between them, and the numbers above the lines are Xoo races against which the epistasis was detected (Tables 3 and 4). Cases for which two interacting loci are connected by solid lines represent the parental type of interaction (the parental genotypes were associated with resistance), those connected by dotted lines represent the recombinant type of interaction (the recombinant genotypes were associated with resistance), and those connected by dotted–dashed lines represent cases for which either the parental or recombinant type interaction was associated with resistance, depending on Xoo races. Rectangular boxes in A and C represent cases for which groups of interacting loci could be included into the networks based on the comparative mapping results in Fig. 1.