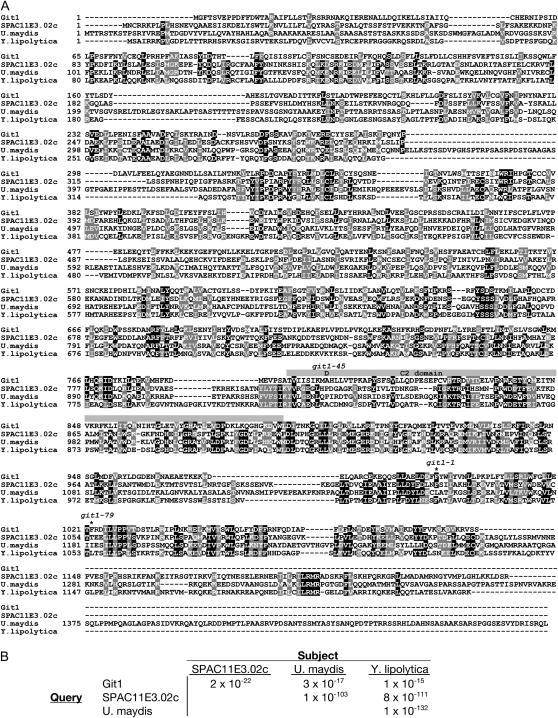
Figure 4.
Alignment of Git1 with SPAC11E3.02c and related proteins from Ustilago maydis and Yarrowia lipolytica. (A) Alignment of S. pombe proteins Git1 (NP_596600) and SPAC11E3.02c (NP_594926) with related proteins from U. maydis (XP_400071) and Y. lipolytica (XP_501312) using the ClustalW sequence alignment program (Thompson et al. 1994) and displayed using BOXSHADE. Identities are shown against a solid background and similarities are shown against a shaded background. A shaded box above the sequences denotes the predicted C2 domain of Git1. The alanine-to-aspartic-acid missense mutation in git1-45 is shown above residue 799 of the Git1 sequence. Asterisks above the Git1 sequence at residues 995 and 1022 denote the last correctly translated residues expressed from alleles git1-1 and git1-79, respectively. (B) E-values associated with alignments from unfiltered BLASTP searches involving the proteins shown in A.