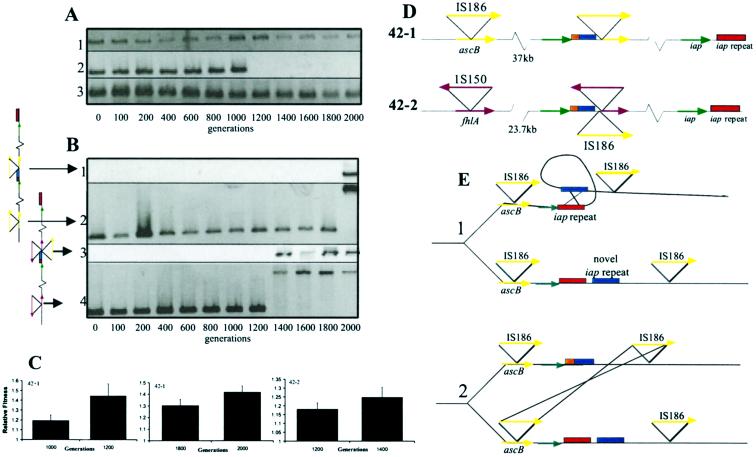
Figure 3.
Confirmation, timing, and molecular characterization of a subset of the duplications and deletions. (A) The 12-kb deletion in the 42+1 line. PCR products for amplifications of sbcB, upstream of the deletion (1.5 kb, row 1), galF, within the deletion (1.1 kb, row 2), and cspG, immediately downstream of the deletion (0.2 kb, row 3), are shown as a function of the number of generations at 41.5°C. (B) Identification of the breakpoints for the tandem duplications in the 42−1 and 42−2 lines. The 37-kb repeat in the 42−1 line occurred between generations 1,800 and 2,000 (middle junction, row 1, 2.5 kb; left breakpoint, row 2, 1.4–2.8 kb). The 23.7-kb repeat in the 42−2 line occurred between generations 1,200 and 1,400 (middle junction, row 3, 4.4 kb; left breakpoint, row 4, 1.1–2.5 kb). Primers used in the amplification of the products in rows 1 and 3 amplify the junction generated by the duplication. Primers used in the amplification of products in rows 2 and 4 amplify the gene at the left breakpoint. The appearance of IS at the left breakpoints is coincident with the origin of the duplications. (C). Relative fitness measurements before and after chromosome rearrangement events. Bars represent means ±95% confidence intervals. (D) Genetic structure of duplications. The unique junction generated by the duplication is represented by the juxtaposition of red and blue blocks. We depict a single duplication, but the number of tandem copies is not determined. (E) Proposed model for the origin of the duplication. The intrastrand recombination is presented in part 1 and the sister strand exchange in part 2. The figure depicts the model for the generation of the 23.7-kb tandem duplication; the model for the 37-kb tandem duplication is identical except that the second recombination event occurs between IS150 located adjacent to the iap repeat sequence and IS150 at the left breakpoint.