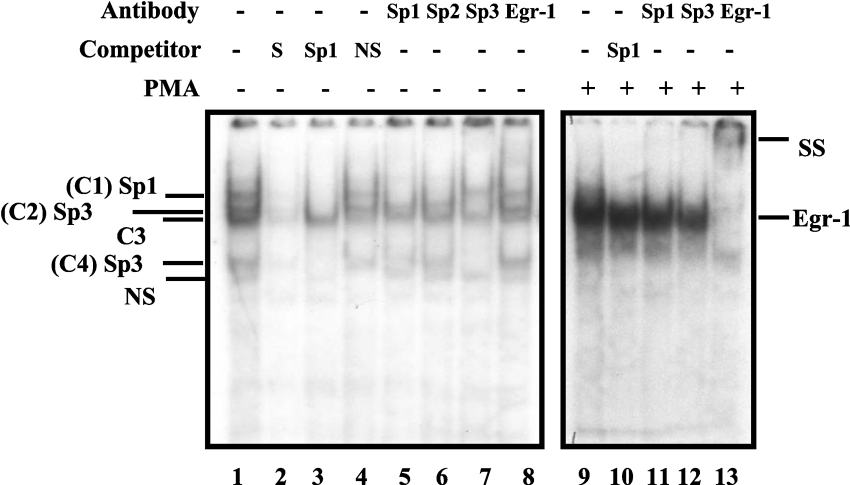
Figure 4. PMA-induced EGR-1 interacts with the downstream EGR-1/Sp1 motif.
GMSA was performed using a double-stranded oligonucleotide (bp −91 to −68) as end-labelled probe and nuclear extracts from untreated or PMA-treated C2BBe1 cells. Nuclear extracts were prepared from C2BBe1 cells at confluence. PMA-treated cells were grown in serum-reduced media at least for 24 h prior to addition of PMA (100 nM) for 2 h. A total of 5 μg of nuclear proteins was combined with 30000 c.p.m. of probe per reaction and, after 20 min incubation at room temperature, resolved on a 5% non-denaturing polyacrylamide gel and visualized by autoradiography. Competition experiments were performed in the presence of unlabelled probe (S), non-specific oligonucleotide (NS) and an oligonucleotide containing Sp1 consensus binding site (Sp1). Supershift assays were performed with anti-Sp1, -Sp2, -Sp3 and -EGR-1 antibodies (2 μg) as indicated. SS denotes the supershifted bands. + and − signs indicate the presence or absence of reaction components in the binding mixture (shown on the top). Protein components of DNA–protein complexes are shown on the right. C1, complex 1.