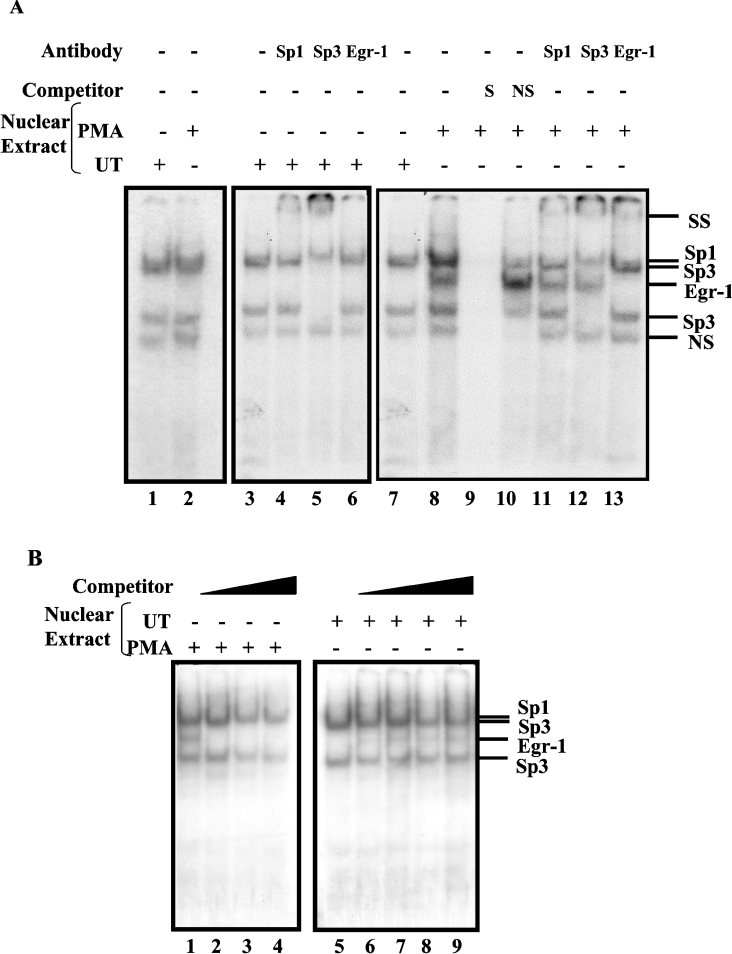
Figure 6. Sp1, Sp3 and EGR-1 interact with the upstream EGR-1/Sp1 motif simultaneously.
(A) GMSAs were performed using 32P-end-labelled probes −270 to −245 (lanes 1 and 2), −148 to −122 (lanes 3–13) with nuclear extracts prepared from the control (lanes 1 and 3–7) and PMA-treated cells (lanes 2 and 8–13). PMA-treated cells were grown in serum-reduced media at least for 24 h prior to addition of PMA (100 nM) for 2 h. The identity of the protein components of the DNA–protein complexes was determined by a competition assay with an unlabelled specific probe (lane 9) and a non-specific oligonucleotide (lane 10) and by supershift assays with anti-Sp1, -Sp3 and -EGR-1 antibodies (lanes 11–13 respectively). DNA–protein complexes were resolved on a 5% native polyacrylamide gel and visualized by autoradiography. (B) Competition assays for protein interactions were performed with −148 to −122 end-labelled probe and PMA-treated nuclear proteins (lanes 1–4) in the presence of 50-, 100- and 200-fold excess of EGR-1 consensus oligonucleotides (lanes 2–4 respectively) or the control nuclear proteins (lanes 5–9) plus increasing amounts of PMA-treated nuclear extracts at 0.5, 1.0, 1.5 and 2.0 μg (lanes 6–9 respectively). The positions of the DNA–protein complexes are shown on the right. SS denotes the supershifted bands. NS, non-specific competitor oligonucleotide or non-specific protein–DNA complex; UT, untreated.