Fig. 1. S. cerevisiae genes containing conserved intragenic repeats.
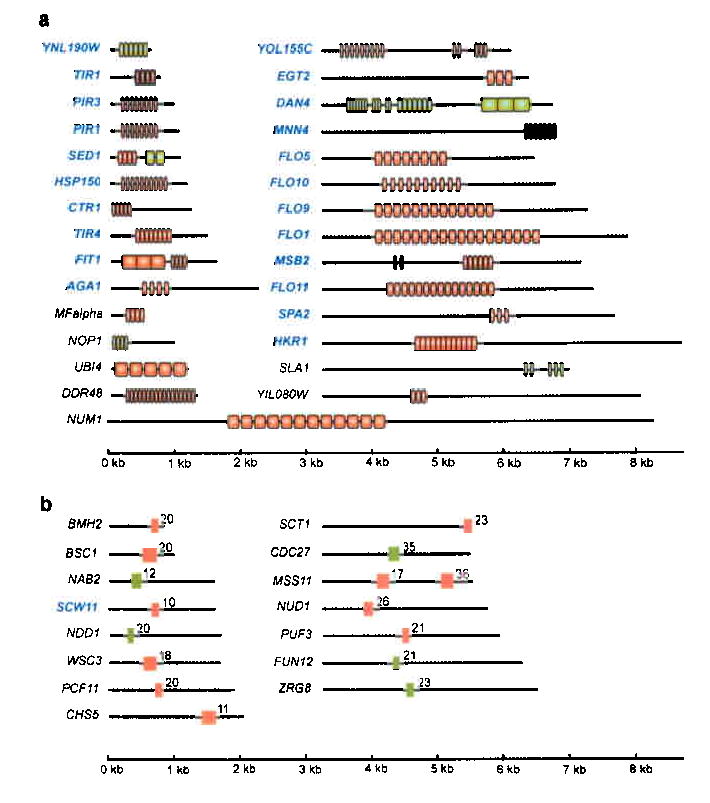
A screen of all open reading frames in the S. cerevisiae genome for those containing conserved intragenic tandem repeats identified 29 genes with large (≥ 40 nt) repeats (panel a) and 15 genes with short (<40 nt) repeats (panel b). Repeats (vertical boxes) that vary in size among 6 different S. cerevisiae strains are colored red (see text and Supplementary Fig. 1-2 online); repeats that do not show size variation among these strains are colored green. Cell surface genes are indicated in blue. The numbers in panel b indicate the number of repeats. More information about the repeats (consensus sequence, conservation…) is given in Supplementary Table 1 online. The repeat units in most genes are distinct from those in others except in FLO1, FLO5 and FLO9, which share the same repeat unit, and in PIR1, PIR3 and HSP150, which share the same repeat unit. SED1, FIT1, SLA1 and MSB2 contain two intragenic repeat regions with different repeat sequences. YOL155C and DAN4 contain three distinct repeat regions.
