Figure 5.
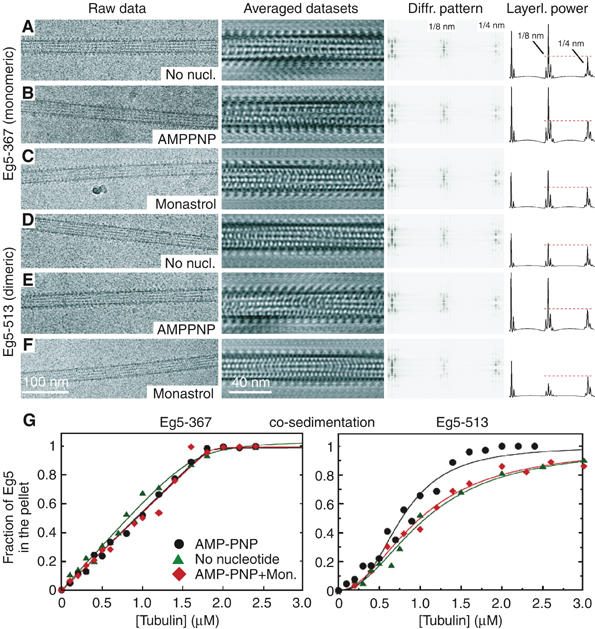
Eg5 motor binding to MTs. Cryo-EM analysis of monomeric MT·Eg5-367 complexes (A–C) and dimeric MT·Eg5-513 complexes (D–F) are shown in different nucleotide states. Diffraction patterns were taken from the averaged data sets. The graphs on the right display the projected total power of each layer line. The leftmost peak is not the equator, but represents the first layer line marking the protofilament supertwist. The 1/4 nm cluster (marked in panel A) relates mainly to the tubulin monomer repeat along the MT axis, whereas 1/8 nm marks the repeat of the αβ-tubulin dimer or a tubulin dimer complexed with one motor head domain. Eg5-367 binds regularly along the MTs in the absence of nucleotides (A), in the presence of MgAMPPNP (B), and in the presence of MgAMPPNP+S-monastrol (C). Dimeric Eg5-513 in the presence of MgAMPPNP also reveals a strong MT affinity (E) that is, however, significantly reduced in the absence of nucleotides (D). AMPPNP+S-monastrol show an even further reduced affinity (F, also see Figure 2E). MT·Eg5 cosedimentation (G) has been performed to compare Eg5-367 (left) and Eg5-513 (right). The MT binding patterns were not significantly different for monomeric Eg5-367 with MgAMPPNP, apyrase-induced nucleotide-free state, or AMPPNP+S-monastrol. In contrast, dimeric Eg5-513 exhibited a sigmoidal MT binding behavior. The AMPPNP state was more tightly bound (KHill=0.49±0.06 μM3, n=2.8±0.3) than the nucleotide-free state (KHill=0.96±0.07 μM2, n=1.7±0.1). Monastrol treatment with AMPPNP (KHill=1.0±0.05 μM2, n=2.0±0.1) resulted in a binding pattern more similar to the apyrase-induced nucleotide-free state.
