Abstract
Wybutosine (yW) is a tricyclic nucleoside with a large side chain found at the 3′-position adjacent to the anticodon of eukaryotic phenylalanine tRNA. yW supports codon recognition by stabilizing codon–anticodon interactions during decoding on the ribosome. To identify genes responsible for yW synthesis from uncharacterized genes of Saccharomyces cerevisiae, we employed a systematic reverse genetic approach combined with mass spectrometry (‘ribonucleome analysis'). Four genes YPL207w, YML005w, YGL050w and YOL141w (named TYW1, TYW2, TYW3 and TYW4, respectively) were essential for yW synthesis. Mass spectrometric analysis of each modification intermediate of yW revealed its sequential biosynthetic pathway. TYW1 is an iron–sulfur (Fe–S) cluster protein responsible for the tricyclic formation. Multistep enzymatic formation of yW from yW-187 could be reconstituted in vitro using recombinant TYW2, TYW3 and TYW4 with S-adenosylmethionine, suggesting that yW synthesis might proceed through sequential reactions in a complex formed by multiple components assembled with the precursor tRNA. This hypothesis is also supported by the fact that plant ortholog is a large fusion protein consisting of TYW2 and TYW3 with the C-terminal domain of TYW4.
Keywords: iron–sulfur cluster, mass spectrometry, ribonucleome analysis, RNA modification, tRNA
Introduction
A characteristic structural feature of functional RNAs is the presence of post-transcriptional modifications. During RNA processing and maturation, RNA molecules undergo various chemical modifications by RNA-modifying enzymes. Accurate maintenance of RNA modifications is required for various biological functions (Björk, 1995; Suzuki, 2005). Our recent studies revealed that lack of tRNA modification leads to translational defects, which have been associated with human diseases (Kirino and Suzuki, 2004; Kirino et al, 2005).
To date, more than 100 different RNA modifications have been reported in RNA molecules from all domains of life (Rozenski et al, 1999). Most have been identified and characterized in tRNA molecules. Many tRNAs have modifications at the first (wobble) position of the anticodon (position 34) and adjacent to the 3′-position of the anticodon (position 37). The wobble modifications at position 34 are important for precise decoding mediated by the codon-anticodon interaction (Suzuki, 2005). Position 37 of the tRNA usually contains a modified purine nucleoside. This modification is often a hyper-modified nucleoside with large molecular weight, such as N6-threonylcarbamoyladenosine (t6A), 2-methylthio-N6-isopentenyladenosine (ms2i6A) or wybutosine (yW). These modifications play a critical role in the stabilization of codon–anticodon pairing through base–stacking interactions and function to maintain the reading frame. yW (whose base is wye, Figure 1A), which contains a tricyclic base, is one of the most complexly modified guanosine residues. yW and its derivatives are found at position 37 of eukaryotic and archaeal phenylalanine tRNA (tRNAPhe) (Figure 1B) (Blobstein et al, 1973), with the exception of Bombyx mori (Keith and Dirheimer, 1980) and Drosophila melanogaster (Altwegg and Kubli, 1979), where 1-methylguanosine (m1G) is present. yW does not have any major influence on the phenylalanylation of tRNAPhe (Thiebe and Zachau, 1968), but may stabilize the first base pair of the codon–anticodon duplex in the ribosomal A site by base stacking (Konevega et al, 2004). tRNAPhe that lacks the yW modification has been shown to enhance frameshifting (Carlson et al, 1999) and influence the replication of human immunodeficiency virus (HIV) RNA (Hatfield et al, 1989). In addition, mouse tRNAPhe isolated from neuroblastoma cells has m1G instead of the naturally occurring hydroxywybutosine (OHyW), and tRNAPhe from Ehrlich ascites tumors contains an undermodified OHyW (OHyW-72; OHyW minus 72 Da) (Kuchino et al, 1982). These data imply that disruption of the yW modification may have pathological consequences.
Figure 1.
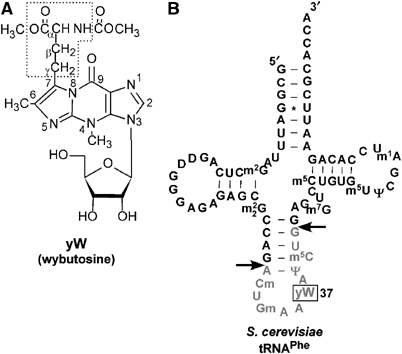
Chemical structure of wybutosine (yW) and secondary structure of tRNAPhe. (A) Chemical structure of yW. Carbon and nitrogen atoms in the tricyclic base are numbered. The α-amino-α-carboxypropyl group at C-7 is boxed by a dotted line. (B) Secondary structure of the S. cerevisiae tRNAPhe with modified nucleosides: wybutosine (yW), 2′-O-methylguanosine (Gm), 2′-O-methylcytidine (Cm), pseudouridine (Ψ), 5-methylcytidine (m5C), 7-methylguanosine (m7G), 2-methylguanosine (m2G), N2,N2-dimethylguanosine (m22G), dihydrouridine (D), 1-methyladenosine (m1A) and 5-methyluridine (m5U). The anticodon-containing fragment produced by RNase T1 digestion is shown in gray. Arrows indicate the sites for RNase T1 cleavage.
The biosynthesis of yW is a multienzymatic process. yW originates from a genetically encoded guanine (Li et al, 1973; Thiebe and Poralla, 1973), and the presence of m1G37 in tRNAsPhe from neuroblastoma cells, as well as B. mori and D. melanogaster, suggested that m1G is the first precursor in the synthesis of yW. N1-methylation of G37 in tRNAPhe is catalyzed by an S-adenosylmethionine(Ado-Met)-dependent tRNA methylase (TRM5) in the initial step of yW synthesis (Droogmans and Grosjean, 1987). Disruption of the TRM5 gene led to a severe growth defect (Björk et al, 2001), suggesting that m1G37 has a critical role in maintaining the correct reading frame in addition to initiating yW synthesis in tRNAPhe. The GAA anticodon of tRNAPhe is necessary for yW synthesis (Droogmans and Grosjean, 1987), although the anticodon is not required for m1G formation by TRM5 (Brule et al, 2004). Thus, subsequent steps in the multienzymatic process from m1G to yW strictly discriminate tRNAPhe from other m1G-containing tRNAs. Very little is known about the biosynthetic steps from m1G to yW. It has been reported that the α-amino-α-carboxypropyl side chain at the C-7 position of the yW-base (Figure 1A) is transferred from methionine or S-adenosylmethionine in yeast cells (Munch and Thiebe, 1975). However, in Vero cells, the lateral chain of the yW-base is derived from lysine (Pergolizzi et al, 1979). These data imply that the biosynthesis of yW in tRNAPhe might vary from one species to another.
Although the number of genes involved in biosynthesis of yW is not known, the genes responsible for yW synthesis must be present among the uncharacterized genes in the Saccharomyces cerevisiae genome. To identify genes responsible for RNA modifications, we have employed a reverse genetic approach combined with mass spectrometry (‘ribonucleome' analysis) (Soma et al, 2003; Suzuki, 2005; Ikeuchi et al, 2006). This analysis utilizes a series of gene-deletion strains of S. cerevisiae. Total RNA extracted from each strain was analyzed by liquid chromatography-mass spectrometry (LC/MS), to determine whether a particular gene deletion leads to the absence of a specific modified base and, thus, permits us to identify the enzyme or protein responsible for this modification. In the case of essential genes, we analyzed temperature-sensitive mutants cultured at the nonpermissive temperature, or expression-controlled strains (Soma et al, 2003). This analysis enables us to identify the enzymes directly responsible for RNA modification, as well as genes that encode nonenzymatic proteins necessary for the biosynthesis of RNA modifications. These include carriers of the metabolic substrates used in the RNA modification and subunit proteins needed for RNA recognition.
Here, using the ribonucleome analysis, we identified four new genes responsible for biosynthesis of yW in S. cerevisiae. Chemical structures of the modification intermediates of yW in these deletion strains enabled us to model a biosynthetic pathway of yW. In addition, yW synthesis was partially reconstituted in vitro. This study will enable us to now develop a complete understanding and in vitro reconstitution of yW synthesis.
Results
Identification of four genes responsible for yW synthesis by ribonucleome analysis
Since yW is a nonessential RNA modification, the complete set of S. cerevisiae deletion strains (4829) serves as a parent population to identify genes responsible for yW synthesis. To reduce the size of the starting population, we selected 3482 genes that have orthologs in Shizosaccharomyces pombe, because S. pombe tRNAs possess all the modified nucleosides found in S. cerevisiae tRNAs. Next, we chose 351 genes with ORFs coding for proteins of unknown function in S. cerevisiae (CYGD: http://mips.gsf.de/genre/proj/yeast) (Guldener et al, 2005) and started the ribonucleome analysis with this population. Parallel small-scale cultivation of each deletion strain was performed in deep 24-well plates. We developed a system that enabled us to obtain total RNA from each cultured strain and perform nuclease digestions in a 96-well format. Samples were then automatically analyzed by LC/MS using ion-trap MS to determine which modified nucleosides, if any, were absent. Our LC/MS analysis routinely detects 20 species of modified nucleosides in yeast total RNA, most are from tRNAs and some are from rRNAs or other noncoding RNAs. In the mass chromatogram, yW is detected as a proton adduct form (MH+) of yWpA (m/z 838, RT 46.8 min), which is a P1 nuclease-resistant dimer (Figure 2A). The protonated base fragment (BH2+) related to yW-base (m/z 377) is also detected (Figure 2A). To search for genes responsible for yW synthesis, we examined mass chromatograms of the 351 selected deletion mutants for peaks at m/z 838 and m/z 377 and identified four deletion strains, YPL207w, YML005w, YGL050w and YOL141w, in which yW was absent (Figure 2A). Since other modified nucleosides in these strains were detected normally (data not shown), the absence of yW is not due to nonspecific reduction of RNA modifications. Moreover, as normal amounts of tRNAPhe could be isolated from each deletion strain, the absence of yW is not due to a specific defect in tRNAPhe transcription (data not shown).
Figure 2.
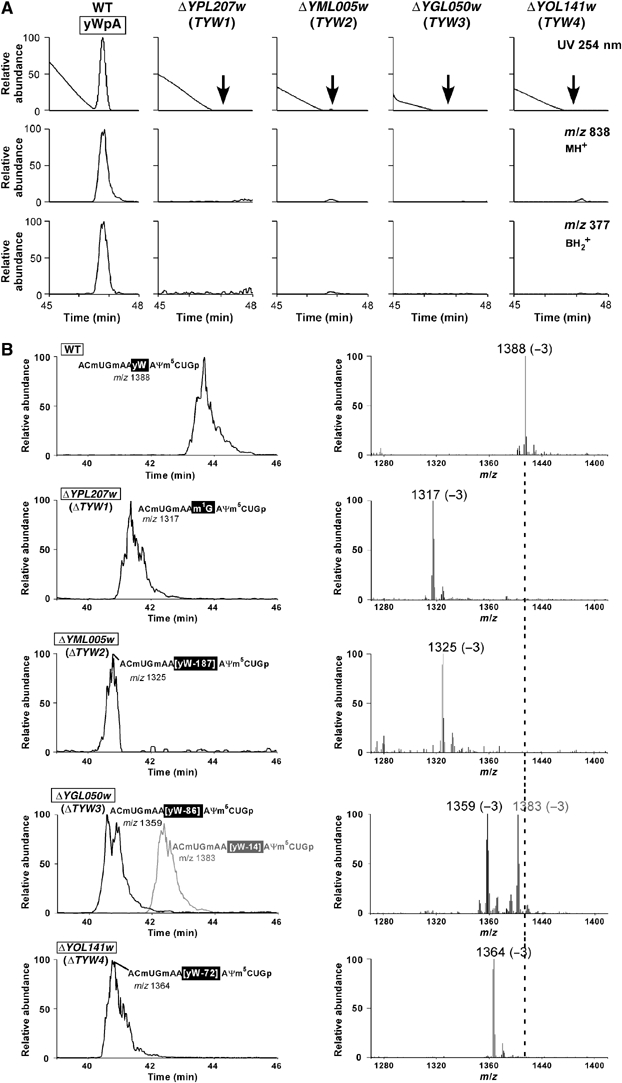
Mass spectrometric analysis of total nucleosides and tRNAPhe from S. cerevisiae wild-type and mutant cells. (A) LC/MS analysis of the total nucleosides in the wild-type (WT), ΔYPL207w (TYW1), ΔYML005w (TYW2), ΔYGL050w (TYW3) and ΔYOL141w (TYW4). The upper panel is the UV trace at 254 nm. The middle and lower panels are mass chromatograms detecting MH+ of yWpA (m/z 838) and BH2+ of yW-base (m/z 377), respectively. Arrows indicate the retention time for yWpA. (B) LC/MS fragment analyses of RNaseT1-digested tRNAPhe obtained from wild-type and ΔTYW1-4. The graphs on the left describe the mass chromatograms shown by triply charged ions of anticodon-containing fragments containing yW (m/z 1388) from WT, m1G (m/z 1317) from ΔYPL207w (TYW1), yW-187 (m/z 1325) from ΔYML005w (TYW2), and yW-86 (m/z 1359) or yW-14 (m/z 1383) from ΔYGL050w (TYW3) and yW-72 (m/z 1364) from ΔYOL141w (TYW4). RNA sequences, including modifications for each fragment, are indicated. The graphs on the right show the mass spectrum for each anticodon-containing fragment. Charge states are indicated in parentheses.
Cell growth of each deletion strain was measured in rich medium as well as in the minimal medium (Supplementary Figure S4). However, we did not observe any significant differences in growth between wild-type and the deletion strains.
Mass spectrometric analysis of the modification intermediates of yW
To confirm the absence of yW at position 37 of tRNAPhe and to determine the chemical structure of each modification intermediate, we isolated individual tRNAPhe from each deletion strain. As expected, the nucleoside analysis revealed that the purified tRNAPhe from each deletion strain had no yW nucleoside (Supplementary Figure S1), but other modified nucleosides were present (data not shown). These results demonstrated that the four new genes encode proteins involved in biosynthesis of yW at position 37 of tRNAPhe. In addition, biosyntheses of other modifications in tRNAPhe (Figure 1B) were not affected by the absence of yW, indicating that yW is not required for biosynthesis of other modifications. We have renamed YPL207w, YML005w, YGL050w and YOL141w as TYW1, TYW2, TYW3 and TYW4 (tRNA-yW synthesizing protein 1–4), respectively.
To determine the mass of each modification intermediate of yW, the purified tRNAPhe was subjected to RNase T1 digestion and analyzed by LC/MS (T1 mapping), as shown in Figure 2B. In the case of wild-type tRNAPhe, the anticodon-containing RNA fragment (ACmUGmAAyWAΨm5CUGp, MW 4167) produced by RNase T1 digestion (Figure 1B) was clearly observed as a triply charged ion with m/z 1388 (Figure 2B). The mass of the same RNA fragment in tRNAPhe obtained from the ΔTYW1 strain was MW 3956 (m/z 1317), which is the mass of a fragment having m1G37 instead of yW37 (Figure 2B). Appearance of m1G was also confirmed by nucleoside analysis of purified tRNAPhe from the ΔTYW1 strain (Supplementary Figure S2A). The data clearly demonstrate that TYW1 encodes a protein responsible for the second step of yW synthesis (see Figure 6). According to T1 mapping of purified tRNAPhe from the other three deletion strains, ΔTYW2, ΔTYW3 and ΔTYW4 generated the modification intermediates yW-187 (yW minus 187 Da, MW 321), yW-86 or yW-14 (yW minus 86 or 14 Da, MW 422 or 494), and yW-72 (yW minus 72 Da, MW 436), respectively, judging from changes in the mass of each anticodon-containing fragment (Figure 2B). These intermediates were also confirmed by nucleoside analysis of the purified tRNAPhe using LC/MS (Supplementary Figure S2B–D). It is intriguing that tRNAPhe from ΔTYW3 strain has two yW intermediates, yW-86 and yW-14. It is likely that yW-86 is a real intermediate at this step and yW-14 is a product by skipping TYW3-dependent reaction. Judging from the intensity of the mass chromatogram, about 50% of tRNAPhe has yW-14 instead of yW-86.
Figure 6.
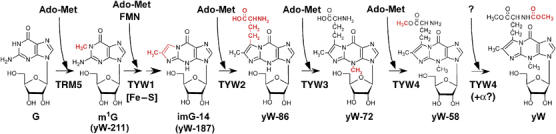
Biosynthetic pathway of yW in S. cerevisiae tRNAPhe. TRM5 methylates G37 of tRNAPhe to produce m1G37, utilizing Ado-Met as a methyl donor. TYW1, an Fe–S cluster protein, may catalyze tricyclic formation of yW using Ado-Met and FMN as cofactors. TYW2 transfers α-amino-α-carboxypropyl group from Ado-Met to the side chain at the C-7 position of the yW-187 to produce yW-86. TYW3 methylates N-4 position of yW-86 in Ado-Met-dependent manner to yield yW-72. Finally, TYW4, an Ado-Met-dependent carboxymethyltransferase, methylates the α-carboxy group of yW-72 to form yW-58, which triggers methoxycarbonylation of α-amino group of yW-58 to complete yW. TYW4 is likely responsible for the last two steps. Substrate for methoxycarbonylation and requirement of partner protein still remain to be investigated.
Chemical structures of the modification intermediates of yW
To further define the chemical structures of the modification intermediates of yW, nucleosides from each purified mutant tRNAPhe were subjected to LC/MS/MS analysis to observe product ions of the bases produced by collision-induced dissociation (CID). The structures of the compounds can then be determined by referring to the reported MS/MS spectra, since characteristic patterns of the product ions are highly correlated to the chemical structure. When yWpA and its intermediates are ionized as proton adducts (MH+) by electrospray ionization (ESI), base fragments of yW and its derivatives are spontaneously generated in the ion source. The protonated base fragment (BH2+) of the each tRNA was selected as a parent mass and decomposed in collision with helium gas to obtain the spectrum of product ions (Supplementary Figure S3). Most of our data could be interpreted by referring to the CID spectra for chemically synthesized yW derivatives, part of the pioneering work by J McCloskey's group (Zhou et al, 2004a, 2004b). Interpretation of each CID spectrum and determination of chemical structure of each yW intermediate are described in the legend for Supplementary Figure S3. The determined chemical structures for yW-187, yW-86, yW-72 and yW-14 are shown in Figure 6 and Supplementary Figure S3.
TYW genes encode highly conserved proteins in eukaryote
Using each yeast TYW gene as a query, we have retrieved homologs from S. pombe, human, mouse and Arabidopsis thaliana. The sequence alignments (Figure 3A, Figure 3B and Figure 3C) show that these proteins share many conserved regions.
Figure 3a.
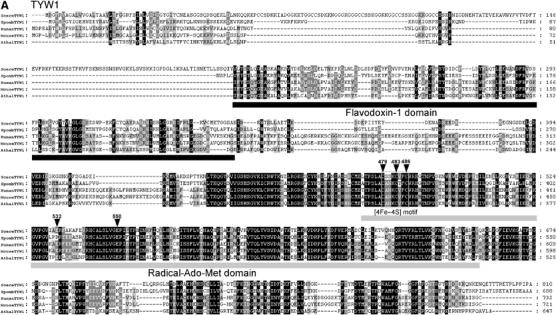
Sequence alignments of the TYW proteins. Each TYW protein is aligned with a set of protein homologs from S. cerevisiae (Scere), S. pombe (Spomb), Homo sapiens (Human), Mus musculus (Mouse), A. thaliana (Athal) and O. sativa (Osati). Multiple alignment of each sequence was carried out by Clustal X (Thompson et al, 1997) and displayed by Genedoc multiple sequence alignment editor (Nicholas et al, 1997). White letters in black boxes represent amino-acid residues identical in all species, while white letters in gray boxes represent residues with ∼80% homology. Black letters in gray boxes represent residues with ∼60% homology. (A) Sequence alignment of TYW1 with its homologs. Two conserved domains, flavodoxin-1 domain and Radical-Ado-Met domain, are underlined. The boxed region represents the [4Fe–4S] motif. Positions for site-directed mutagenesis are indicated by arrowheads.
Figure 3b.

(B) Sequence alignment of TYW2 and TYW3 with its homologs. A. thaliana and O. sativa homologs are large fusion proteins including TYW3 and the C-terminal region of TYW4 and TYW2 (TYW3-4C-2). A set of TYW2 homologs and TYW3 homologs are aligned with plant TYW3-4C-2. The Met-10+-like protein family domain in TYW2 is indicated by a line. The C-terminal region of TYW4 in plant TYW3-4C-2 is shaded.
Figure 3c.

(C) Sequence alignment of TYW4 with its homologs. The conserved domain, LCM, is indicated by a line. The boxed regions are Ado-Met-binding motifs, including motif I–III and post-I. The residues important for protein carboxyl methyltransferase activity are indicated by arrowheads. The C-terminal region of TYW4 is shaded. (D) Domain structure of the plant TYW3-4C-2 protein. The plant TYW3-4C-2 protein is a large fusion protein composed of TYW3, the CTD of TYW4 and TYW2.
TYW1 (810 aa in S. cerevisiae) and its homologs contain two conserved domains (Figure 3A), a flavodoxin-1 domain in the N-terminus (pfam00258) and a radical-Ado-Met domain in the C-terminus (pfam04055). Flavodoxin is a protein that includes an iron–sulfur (Fe–S) cluster and employs FMN as a cofactor (Wakabayashi et al, 1989). Radical Ado-Met proteins contain a conserved CxxxCxxC motif (Figure 3A), which is responsible for binding a [4Fe–4S] cluster, as a catalytic core for diverse reactions (Sofia et al, 2001). These structural features of the conserved domains suggest that TYW1 is an Fe–S cluster protein responsible for ring formation of the tricyclic base of yW.
TYW2 (462 aa in S. cerevisiae) and its homologs contain a conserved Ado-Met-binding motif found in Met-10+-like protein family (lower part of Figure 3B) (Niewmierzycka and Clarke, 1999). In addition, TYW2 was found to be a member of COG2520, which also includes TRM5. The data indicate that TYW2 is an Ado-Met-dependent enzyme.
TYW3 (273 aa in S. cerevisiae) was found to be a highly conserved protein (pfam02676/DUF207, COG1590), but its function is unknown. A. thaliana and Oryza sativa homologs of TYW3 have a long extended C-terminal domain (CTD) of about 1000 aa (Figure 3B), and it is interesting to note that the long extended CTD of the plant TYW3 actually contains the CTD of TYW4 as well as the entire sequence of TYW2. Thus, the plant homologs appear to be a large fusion protein including TYW3, the CTD of TYW4 and TYW2, which we have named TYW3-4C-2 (Figure 3B and D). Additionally, the plant TYW4 is a short protein without the CTD (Figure 3C). These data suggest that TYW3-4C-2 acts as a multienzymatic complex for yW synthesis in A. thaliana and O. sativa. Schematic depiction of the topological orientation of TYW2, TYW3, TYW4 and TYW3-4C-2 is shown in Figure 3D.
TYW4 (695 aa in S. cerevisiae) has a conserved leucine carboxyl methyltransferase (LCM) domain (pfam04072) in its N-terminal region (Figure 3C). TYW4 has an apparent paralog, PPM1 (protein phosphatase methyltransferase 1), which is a yeast methyltransferase responsible for methylesterification of the C-terminal Leu residue of PP2A (protein phosphatase 2A) (Wu et al, 2000; Kalhor et al, 2001). Thus, YOL141w (TYW4) had been previously named PPM2 (Kalhor et al, 2001; Wei et al, 2001), but its biochemical function was never demonstrated directly. The PPM family has three highly conserved Ado-Met-binding motifs (motifs I, II and III). This finding strongly suggests that, using Ado-Met as a methyl donor, TYW4 catalyzes methylation of the carboxyl group of the methionine moiety in yW-72 to convert it to yW-58 (see Figure 6).
Biogenesis of an Fe–S cluster is required for TYW1 function in yW synthesis
The S. cerevisiae NFS1 gene encodes a cysteine desulfurase involved in Fe–S cluster biogenesis. To test whether yW synthesis requires biosynthesis of an Fe–S cluster, we analyzed total nucleosides from S. cerevisiae YN101 cells in which expression of NFS1 is controlled under the GAL1 promoter (Nakai et al, 2004). We previously demonstrated that inhibition of NFS1 expression resulted in a severe reduction of thio-modifications in both mitochondrial and cytoplasmic tRNAs (Nakai et al, 2004). When cells were cultured in galactose-containing medium, yWpA was clearly observed (Figure 4A). However, when the expression of NFS1 was downregulated by culturing cells in glucose-containing medium, yWpA was severely reduced (Figure 4A). These data demonstrated that biosynthesis of Fe–S clusters by NFS1 is absolutely required for yW synthesis. The Fe–S cluster in TYW1 is likely to be critical for the formation of the tricyclic base of yW.
Figure 4.
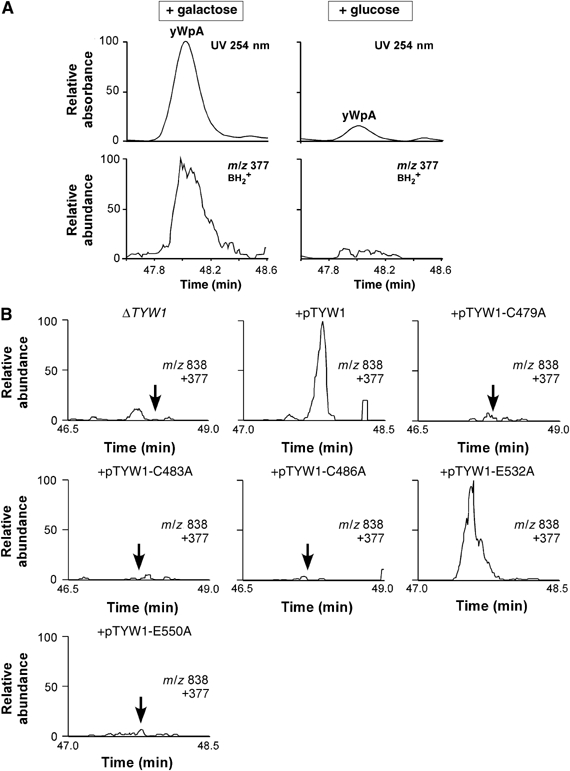
Fe–S cluster in TYW1 involved in yW synthesis. (A) LC/MS analysis of the total nucleosides in the YN101 cells in which expression of NFS1 is controlled under the GAL1 promoter. The left- and right-hand graphs describe the LC/MS chromatograms of yWpA from YN101 cultured with galactose and glucose, respectively. The upper panels are the UV trace at 254 nm. The lower panels are mass chromatograms detecting the yW-base (m/z 377). The relative amount of yWpA in both conditions was normalized to the pseudouridine (Ψ) content. (B) Complementation test of ΔTYW1 by introducing a series of mutant plasmid pTYW1 in which critical residue for Fe–S cluster is mutated. Total nucleosides from each strain were analyzed by LC/MS to detect yWpA. Merged mass chromatograms of yWpA (m/z 838) and yW-base (m/z 377) are shown. Arrows indicate the retention time for yWpA. yWpA was restored by introducing wild-type pTYW1 or pTYW1-E532A. Other mutant plasmids did not complement yW synthesis in ΔTYW1 strain.
The radical-Ado-Met domain in the C-terminal region of TYW1 has a conserved C479xxxC483xxC486 motif (Figure 3A) for [4Fe–4S] cluster binding. In addition, E532 and E550 are candidate residues for GGE motif in the radical Ado-Met domain (Pierrel et al, 2004). To clarify functional importance of these conserved residues, site-directed mutagenesis of Cys479, Cys483, Cys486, Glu532 and Glu550 was carried out. The plasmid expressing TYW1 (pTYW1) with each mutation was introduced into ΔTYW1 strain to test complementation activity of each mutant for yW synthesis. As shown in Figure 4B, yW in the ΔTYW1 strain was restored by the introduction of wild-type pTYW1. No activity for yW synthesis could be observed when pTYW1 with C479A, C483A, C486A or E550A mutation was introduced. E532A mutation did not affect TYW1 functionality. These results revealed that three conserved Cys in the CxxxCxxC motif as well as E550 in the GGE motif is critical for yW synthesis. Taken together with the result of NFS1 data, biogenesis of an Fe–S cluster in TYW1 is required for TYW1 function in yW synthesis.
Reconstitution of yW synthesis in vitro using recombinant TYW2, TYW3 and TYW4
Next, we performed in vitro reconstitution of yW synthesis using recombinant TYW proteins. A hexahistidine-tagged TYW2, TYW3 and TYW4 were expressed in S. cerevisiae and purified using an Ni+-chelating column. tRNAPhe having yW intermediate purified from each deletion strain was employed as a substrate for each step of yW synthesis in vitro.
In vitro reconstitution of the tRNAPhe having yW-187 by recombinant TYW2 was carried out in the presence or absence of Ado-Met. As shown in Figure 5A, in the absence of Ado-Met, the substrate tRNA yielded an anticodon-containing RNA fragment whose mass is unchanged (ACmUGmAA(yW-187)AΨm5CUGp, MW 3980). In the presence of Ado-Met, yW-187 in tRNAPhe was clearly converted to yW-86 (101 Da increase). No other product, such as yW-173 (14 Da increase), was detected. The data revealed that recombinant TYW2 has an activity to transfer α-amino-α-carboxypropyl group from Ado-Met to the side chain at the C-7 position of the yW-base. This result is consistent with a previous observation that α-amino-α-carboxypropyl group of yW originates from methionine or Ado-Met in yeast cells (Munch and Thiebe, 1975).
Figure 5.
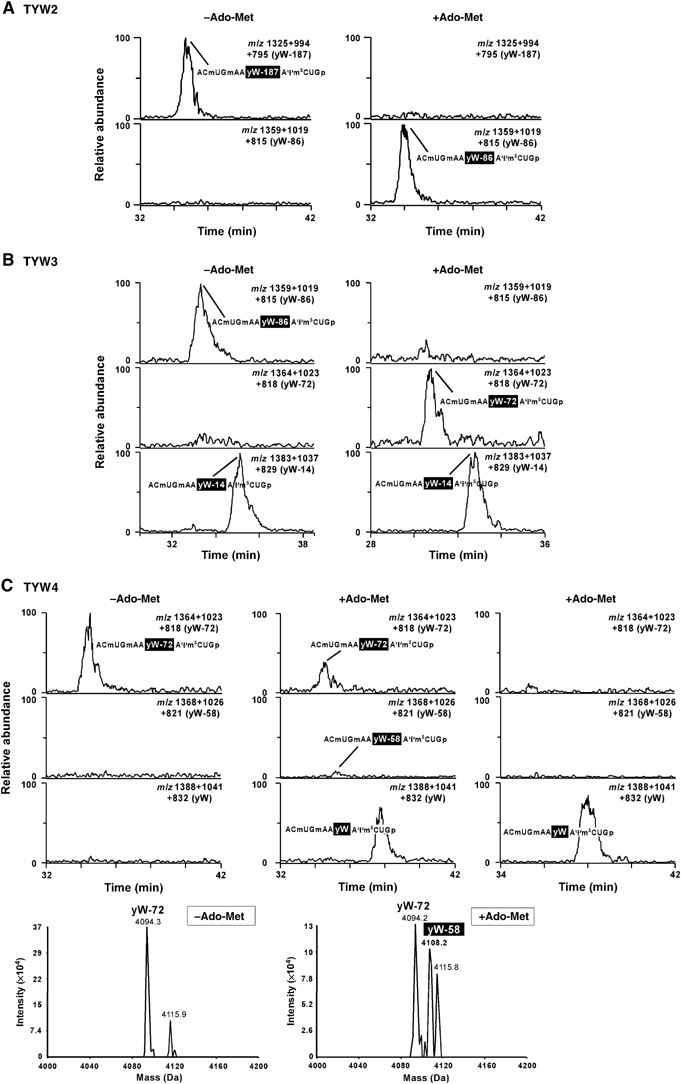
In vitro reconstitution of yW synthesis using recombinant TYW2, TYW3 and TYW4. LC/MS fragment analysis of RNaseT1-digested tRNAPhe whose yW is partially reconstituted by recombinant TYW2 (A), TYW3 (B) and TYW4 (C) in the presence or absence of Ado-Met. All mass chromatograms are produced by integration of triply (−3), quadruply (−4) and quintuply (−5) charged ions of anticodon-containing fragments. (A) tRNAPhe having yW-187 treated by recombinant TYW2 in the absence (left panels) or in the presence (right panels) of Ado-Met. The top and bottom graphs show mass chromatographs for anticodon-containing fragments containing yW-187 (m/z 1325+994+795) and yW-86 (m/z 1359+1019+815), respectively. (B) tRNAPhe having yW-86 (and yW-14) treated by recombinant TYW3 in the absence (left panels) or in the presence (right panels) of Ado-Met. The top, middle and bottom graphs show mass chromatographs for anticodon-containing fragments containing yW-86 (m/z 1359+1019+815), yW-72 (m/z 1364+1023+818) and yW-14 (m/z 1383+1037+829), respectively. (C) tRNAPhe having yW-72 treated by recombinant TYW4 in the absence (left panels) or in the presence (middle and right panels) of Ado-Met. Experiment in the right panels was carried out by highly purified recombinant TYW4. The top, middle and bottom graphs show mass chromatographs for anticodon-containing fragments containing yW-72 (m/z 1364+1023+818), yW-58 (m/z 1368+1026+821) and yW (m/z 1388+1041+832), respectively. Deconvolution spectra of the mass spectra (RT 34–36 min) for the anticodon-containing fragments having yW-72 reconstituted in the absence (left bottom) or presence (right bottom) of Ado-Met. The fragment with yW-58 was clearly detected in the presence of Ado-Met.
In ΔTYW3 strain, tRNAPhe has two yW intermediates, yW-86 and yW-14 (Figure 2B). As shown in Figure 5B, recombinant TYW3 clearly methylated yW-86 to yield yW-72 in Ado-Met-dependent manner. Thus, TYW3 is an N-4 methyltransferase for yW synthesis. On the other hand, yW-14 was not methylated by TYW3 (no yW product is obtained). This suggests that complete synthesis of the side chain at C-7 position inhibits N-4 methylation by TYW3.
The strong homology between TYW4 and PPM1 prompted us to test the evolutionary link between RNA methylase and protein methylase. tRNAPhe having yW-72 purified from ΔTYW4 cells was employed as a substrate for the yW reconstitution. In the presence of Ado-Met (Figure 5C, middle panels), UV trace of the chromatogram showed that about 60% of the anticodon-containing RNA fragment with yW-72 was converted to yield the fragments with yW-58 and yW in which the mass had increased by 14 and 72 Da, respectively. We could detect scanty amount of the RNA fragment with yW-58, which is expected as a direct product of TYW4 methylation, by the deconvolution analysis of the mass spectrum (Figure 5C). The data clearly demonstrated that recombinant TYW4 has an apparent Ado-Met-dependent methylation activity. However, it was unexpected that yW was a major product by TYW4-catalyzed reconstitution of yW-72. As RNA fragments with yW-14 or other possible intermediates were not observed (Figure 5C), these results indicate that multistep enzymatic formation of yW from yW-72 is triggered by Ado-Met-dependent TYW4 methylation. We thought small quantity of other protein involved in methoxycarbonylation of yW-58 might be copurified with the his-tagged TYW4 protein, since the recombinant TYW4 was obtained by partial purification by an Ni+-chelating column. Then, the recombinant TYW4 protein was subjected to further purification by anion exchange chromatography. In vitro reconstitution of yW-72 was then performed using the purified TYW4. As shown in the right panels in Figure 5C, almost all yW-72 was directly converted to yW without showing any yW-58 peak. The result strongly suggests that TYW4 is a bifunctional enzyme to synthesize yW by catalyzing methylation and methoxycarbonylation of yW-72.
Discussion
Biosynthesis of yW is a multienzymatic process, and to determine the precise order of each gene in the biosynthetic pathway of yW, we analyzed the chemical structures of the yW intermediates produced by each deletion strain. In the ΔTYW1 strain, the modification intermediate was m1G. Since m1G is the first intermediate for yW synthesis catalyzed by TRM5, TYW1 therefore encodes an enzyme responsible for the second step of the reaction employing m1G37-containing tRNAPhe as a substrate. The chemical structure of the reaction product is unknown. The fact that a TYW1 homolog is not encoded in the Drosophila genome is consistent with the observation that insect tRNAPhe utilizes m1G37 instead of yW37 (Altwegg and Kubli, 1979; Keith and Dirheimer, 1980). In addition, mouse tRNAPhe from neuroblastoma cells have m1G37 instead of OHyW37 (Kuchino et al, 1982), suggesting that a functional defect in the mouse homolog of TYW1 or its partner proteins might occur in these cells. TYW1 and its homologs share the flavodoxin-like domain and the radical Ado-Met domain (Figure 3A). The flavodoxin-like domain is found in a number of proteins including flavodoxin and nitric oxide synthase. Flavodoxin is found in electron-transfer proteins that function in various electron transport systems. These domains bind one FMN molecule, which serves as a redox-active prosthetic group and are functionally interchangeable with ferredoxins (Wakabayashi et al, 1989). Radical Ado-Met proteins catalyzes diverse reactions, including methylation, isomerization, sulfur insertion, ring formation, anaerobic oxidation and protein radical formation (Sofia et al, 2001). The conserved CxxxCxxC motif ([4Fe–4S] motif in Figure 3A) in the radical Ado-Met proteins is responsible for binding the [4Fe–4S] cluster, which is involved in the reductive cleavage of Ado-Met to generate the 5′ deoxyadenosyl radical (Ado°) (Fontecave et al, 2001; Frey and Booker, 2001). Ado° initiates radical-based chemistry on various substrates. MiaB, which is responsible for biosynthesis of ms2i6A, is shown to be a radical Ado-Met enzyme involved in methylthiolation of isopentenyl-adenosine (i6A) at position 37 of some tRNAs (Pierrel et al, 2004). In this study, inhibition of NFS1 expression resulted in severe reduction of yW. Three conserved Cys residue and Glu550 in the motifs were essential for TYW1 function. These results clearly demonstrated that the involvement of Fe–S clusters in yW synthesis. In the crystal structure of RumA, which is an Fe–S cluster-containing protein responsible for rRNA methylation, its Fe–S cluster domain is directly involved in RNA recognition (Lee et al, 2005). It can be speculated that Fe–S cluster in TYW1 might have an important role in tRNA recognition and ring formation of tricyclic base of yW. To elucidate the detailed mechanism of this reaction, it is necessary to determine the carbon source of C-6 and its methyl group in the tricyclic base.
yW-187 (imG-14) was the yW intermediate obtained from the ΔTYW2 strain. TYW2 has an Ado-Met-dependent methyltransferase motif found in the Met-10+-like protein family, implying that TYW2 catalyzes the N-4 methylation of imG-14 to produce imG (wyosine) using Ado-Met as a methyl donor. imG has an isomer named imG2 (isowyosine) (Blobstein et al, 1973; Kasai et al, 1976), which has a methyl group at C-7 of imG-14 and there was another possibility that TYW2 methylates the C-7 position of imG-14 to produce imG2. However, according to our in vitro reconstitution, TYW2 has an activity to transfer α-amino-α-carboxypropyl group from Ado-Met to the side chain at the C-7 position of the yW to synthesize yW-86. Recently, Kalhor et al (2005) reported that deletion of YML005w (also called TRM12) generates imG-14, although the enzymatic activity was not directly demonstrated.
yW-86 was identified to be an yW intermediate in the ΔTYW3 strain. Recombinant TYW3 was found to be N-4 methyltransferase that methylates yW-86 to yield yW-72 in Ado-Met-dependent manner. In addition, yW-14, which lacks N-4 methyl group from yW, was also found in this strain, suggesting that N-4 methyl group is not essential for complete synthesis of the C-7 side chain mediated by TYW4. Since yW-14 was never a substrate for TYW3, it is considered that complete C-7 side chain inhibits N-4 methylation by TYW3.
yW-72, found in the ΔTYW4 strain, is a yW intermediate, which has an α-amino-α-carboxypropyl group at C-7 of the tricyclic base (Figure 6). TYW4 (previously named PPM2) was expected to be an Ado-Met-dependent methyltransferase having a conserved LCM domain also present in the protein methyltransferase PPM1 (Wu et al, 2000; Kalhor et al, 2001). In this study, yW-72 of tRNAPhe was methylated in an Ado-Met-dependent manner in the presence of TYW4 in vitro, demonstrating that TYW4 is a bona fide methyltransferase for yW-72. The N-terminal domain of TYW4/PPM2 is homologous to PPM1. Considering that PPM1 methylates the C-terminal carboxyl group of two PP2A proteins (PPH21 and PPH22) (Wu et al, 2000; Kalhor et al, 2001), TYM4/PPM2 should methylate the carboxyl group of the Met moiety of yW-72. The observation that PPM2 has no methylation activity for PP2A proteins (Kalhor et al, 2001) is consistent with our findings in this study. The crystal structure of PPM1 complexed with Ado-Met showed a common Ado-Met-dependent methyltransferase fold with a cavity for binding the PP2A C-terminal peptide (Leulliot et al, 2004), where C202, Y206, N250, L251 and R255 of PPM1 were shown to be important for protein methylation (Leulliot et al, 2004). In the case of TYW4, the surface of the cavity is composed of TYW4-specific residues, such as V225, H272 and F273, in the flexible α5 helix (Figure 3C). Since the flexibility of the α5 helix is involved in substrate binding, the differences in this region may recognize yW-72-containing tRNAPhe. TYW4 has a specific extension of the CTD, which is not present in PPM1. The TYW4-specific CTD is likely to be involved in recognition of tRNAPhe. The presence of the TYW4-specific CTD in the large fusion protein TYW3-4C-2 in Arabidopsis also supports the importance of this domain for yW synthesis. The high sequence similarity suggests that TYW4 may have evolved from the PPM1-like carboxyl methyltransferase by acquiring the CTD, which is responsible for recognition of the RNA molecule.
The plant homologs of TYW2 and TYW3 are part of a large fusion protein that we have named TYW3-4C-2, which includes the CTD of TYW4 (Figure 3B and D). It is interesting that the absence of the CTD in plant TYW4 is explained by the presence of a TYW4-CTD in TYW3-4C-2. As there is one TYW4 (or PPM1) homolog in each plant genome, plant TYW4/PPM1 might be a bifunctional protein responsible for methylation of the C-terminal carboxyl group of PP2A protein and methylation of yW intermediates. The topological orientation of TYW3-4C-2 in plant prompted us to speculate that yW synthesis proceeds through consecutive sequential reactions in a multienzyme complex. If TYW4-CTD is responsible for recognition of tRNAPhe, plant TYW4 might coassemble with the tRNAPhe–TYW3-4C-2 complex (Figure 7). In S. cerevisiae or in mammals, TYW2, TYW3 and TYW4 might assemble with tRNAPhe in an analogous manner to carry out the multistep reactions in yW synthesis (Figure 7). Since TYW proteins were not identified as interacting partners of other TYW proteins in the S. cerevisiae protein–protein network database (Supplementary Table S1), it is possible that tRNAPhe serves as a scaffold to form this multienzymatic complex. Other unidentified proteins involved in yW synthesis might be found in the tRNAPhe-dependent complex. In general, assembly of several proteins responsible for multistep biosynthesis with a substrate enables sequential reactions to proceed more smoothly, due to the close proximity of enzymes and substrates within the complex.
Figure 7.
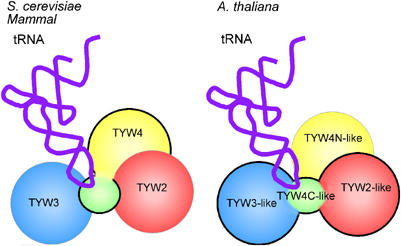
Schematic depiction of a multienzymatic complex of TYW proteins. In S. cerevisiae and in mammals, TYW2, 3 and 4 might coassemble with tRNAPhe to carry out the sequential reactions of yW synthesis. In plants, a large fusion protein of TYW3-4C-2 and TYW4 might coassemble with tRNAPhe.
The predicted biosynthetic pathway of yW is depicted in Figure 6. First, the N1-methyltransferase TRM5 methylates G37 of tRNAPhe to convert it to m1G37. Second, m1G37 serves as a substrate for the Fe–S cluster protein TYW1, to form the tricyclic core of yW, using FMN as a cofactor. Next, TYW2 transfers α-amino-α-carboxypropyl group from Ado-Met to the lateral side chain at the C-7 position of the yW-187 to form yW-86. TYW3 catalyzes N-4 methylation of yW-86 to yield yW-72 in Ado-Met-dependent manner. Finally, TYW4, an Ado-Met-dependent carboxymethyltransferase, methylates the α-carboxy group of yW-72 to form yW-58. Presumably, second function of TYW4 or involvement of unidentified factor(s) should be responsible for methoxycarbonylation of the α-amino group of the side chain of yW-58 to form yW. This multistep enzymatic formation from yW-72 to yW is triggered by TYW4 methylation in a tRNAPhe-dependent protein complex.
Localization of many yeast ORFs has been investigated (Kumar et al, 2002; Huh et al, 2003; Sickmann et al, 2003) and the localization of each TYW protein is listed in Supplementary Table S1. TRM5 localizes to the nucleus and cytoplasm (Huh et al, 2003) and the initial step of yW synthesis could occur in the nucleus. However, other tRNA-processing enzymes localize to the mitochondrial outer membrane (Yoshihisa et al, 2003). More recently, active shuttling of tRNA between the nucleus and cytosol in yeast has been reported (Takano et al, 2005). Thus, localization of the first step of yW synthesis remains an open question. TYW1 localizes on the endoplasmic reticulum (ER). TYW2 localizes to the cytoplasm, ER and nuclear envelope, and TYW4 localizes to the cytoplasm and mitochondria. In addition, the predicted localization of the A. thaliana TYW3-4C-2 is cytoplasmic (http://www.arabidopsis.org/). These data suggest that the subsequent step of yW synthesis takes place mainly in the cytoplasm. Post-transcriptional modifications serve as signals that control the subcellular localization of eukaryotic RNA molecules (Kaneko et al, 2003; Suzuki, 2005). In future studies, biosynthesis of yW should be investigated in association with subcellular localization of a precursor tRNAPhe having yW intermediates with different chemical structures.
Materials and methods
Strains and media
S. cerevisiae wild-type strain and deletion strains were obtained from EUROSCARF: the BY4742 (Matα; his3Δ1; leu2Δ0; lys2Δ0; ura3Δ0) series and strains Y14418, Y10571, Y16650 and Y11085 (BY4742, YGL050w∷kanmx4, YML005w∷kanmx4, YOL141w∷kanmx4, YPL207w∷kanmx4). For recombinant protein production, the S. cerevisiae strain BY4741 (Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) overexpressing hexahistidine-tagged YML005w (YSC3869-9514898), YGL050w (YSC3869-9514409) and YOL141w (YSC3869-9515336) were obtained from Open Biosystems. The BG1805-derived plasmid containing the YPL207w (named pTYW1) was obtained from Open Biosystems. Strains were grown in rich medium (YPD, 2% peptone, 1% yeast extract and 2% glucose), in minimal medium (SD, 0.67% yeast nitrogen base without amino acids and 2% glucose) or in synthetic complete medium (SC, 0.67% yeast nitrogen base without amino acids, 0.5% casamino acid and 2% glucose) supplemented by auxotrophic nutrients as specified. YN101 was cultured as described (Nakai et al, 2004).
Ribonucleome analysis
Parallel preparation of total RNAs from yeast strains. Yeast strains were grown in 5 ml of YPD in a 24-well format deep well plates at 30°C for 36 h and cells were harvested during log phase growth (OD660≈1.5–2.0). Cell pellets were resuspended in 500 μl of lysis buffer (20 mM Tris–HCl (pH 7.5), 10 mM MgCl2), and extracted with neutral pH phenol/chloroform (500/100 μl), which was added to the lysate and shaken for 3 h at room temperature. RNA was then recovered by ethanol precipitation from the aqueous phase. The total RNA samples were dissolved in 100 μl of ddH2O and stored at −20°C.
Mass spectrometry. To analyze RNA nucleosides, total RNA (20 μg) obtained from each strain was digested to nucleosides with nuclease P1(Yamasa) and bacterial alkaline phosphatase derived from Escherichia coli strain C75 (BAP.C75, Takara) for 3 h at 37°C, and analyzed by LC/MS using ion-trap MS as described previously (Kaneko et al, 2003) with slight modifications. Nucleosides were separated by an ODS reverse-phase column (Intertsil ODS3 5 μm, 2.1 × 250 mm, GL Science) using an HP1100 liquid chromatography system (Agilent). The solvent consisted of 0.1% acetonitrile in 5 mM NH4OAc (pH 5.3) (Solvent A) and 60% acetonitrile in H2O (Solvent B) in the following gradients: 1–35% B in 0–35 min, 35–99% B in 35–40 min, 99% B in 40–50 min, 99–1% B in 50–50.1 min and 1% B in 50.1–60 min. The chromatographic effluent was directly conducted to the ESI source to ionize the separated nucleosides, which were analyzed on an LCQ DUO ion-trap mass spectrometer (ThermoElectron). The mass spectrometer was operated with a spray voltage of 5 kV and a capillary temperature of 245°C. The sheath gas flow rate was 95 arb, auxiliary gas flow rate was 5 arb. Positive ions were scanned over an m/z range of 100–900.
T1 mapping and CID
RNA fragments produced by RNase T1 digestion were analyzed by LC/MS using ion-trap MS according to previously described methods (Kaneko et al, 2003; Soma et al, 2003) with slight modifications. Purified tRNAPhe (2.5 μg) was subjected to digestion by RNase T1 (Seikagaku corporation; 0.6 U) for 30 min at 37°C in a buffer of 10 mM NH4OAc (pH 5.3). The hydrolysates were analyzed by LC/MS using a ODS column (Xterra MS C18 2.5 μm, 2.1 × 50 mm, Waters). The solvent consisted of 0.4 M 1,1,1,3,3,3-hexafluoro-2-propanol (pH 7.0, adjusted with triethylamine) in H2O (Solvent A) and 50% methanol (Solvent B) in the following gradients: 5% B in 0–5 min, 5–95% B in 5–35 min, 95% B in 35–40 min, 95–5% B in 40–42 min and 5% B in 42–60 min. Negative ions were scanned over an m/z range of 620–2000 throughout the separation. LC/MS/MS analysis for the yW-base and its derivatives was performed by ion-trap mass spectrometry using the LCQ DUO. The collision energy was 25–30%. Product ions were scanned over an m/z range of 100–900.
In vitro reconstitution of yW synthesis using recombinant proteins
Reaction mixtures of 10 μl containing 50 mM Tris–HCl (pH 8.0), 0.5 mM DTT, 10 mM MgCl2, 1 mM spermidine, 2 μg of tRNAPhe having yW intermediate obtained from each deletion strain, and 1.4 μM recombinant TYW2, TYW3 or TYW4 protein, with or without 0.5 mM Ado-Met, were incubated 1 h at 30°C. After incubation, the reaction was stopped by adding 0.5 M Tris–HCl (pH 8.0) and phenol. The tRNAPhe was extracted and precipitated by ethanol and then subjected to RNase T1 digestion. The RNA fragment analysis by LC/MS was carried out as described above.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to Takeo Suzuki and Noriko Umeda (University of Tokyo) for technical supports, and Yumi Nakai (Osaka Medical College) for providing material. This work was supported by grant-in-aid for scientific research on priority areas from the Ministry of Education, Science, Sports and Culture of Japan (to TS) and by a grant from the New Energy and Industrial Technology Development Organization (NEDO) (to TS).
References
- Altwegg M, Kubli E (1979) The nucleotide sequence of phenylalanine tRNA2 of Drosophila melanogaster: four isoacceptors with one basic sequence. Nucleic Acids Res 7: 93–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR (1995) Biosynthesis and function of modified nucleosides. In Söll D, Rajbhandary UL (eds). tRNA: Structure, Biosynthesis, and Function, pp 165–205. Washington, DC: ASM Press [Google Scholar]
- Björk GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP (2001) A primordial tRNA modification required for the evolution of life? EMBO J 20: 231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobstein SH, Grunberger D, Weinstein IB, Nakanishi K (1973) Isolation and structure determination of the fluorescent base from bovine liver phenylalanine transfer ribonucleic acid. Biochemistry 12: 188–193 [DOI] [PubMed] [Google Scholar]
- Brule H, Elliott M, Redlak M, Zehner ZE, Holmes WM (2004) Isolation and characterization of the human tRNA-(N1G37) methyltransferase (TRM5) and comparison to the Escherichia coli TrmD protein. Biochemistry 43: 9243–9255 [DOI] [PubMed] [Google Scholar]
- Carlson BA, Kwon SY, Chamorro M, Oroszlan S, Hatfield DL, Lee BJ (1999) Transfer RNA modification status influences retroviral ribosomal frameshifting. Virology 255: 2–8 [DOI] [PubMed] [Google Scholar]
- Droogmans L, Grosjean H (1987) Enzymatic conversion of guanosine 3′ adjacent to the anticodon of yeast tRNAPhe to N1-methylguanosine and the wye nucleoside: dependence on the anticodon sequence. EMBO J 6: 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontecave M, Mulliez E, Ollagnier-de-Choudens S (2001) Adenosylmethionine as a source of 5′-deoxyadenosyl radicals. Curr Opin Chem Biol 5: 506–511 [DOI] [PubMed] [Google Scholar]
- Frey PA, Booker SJ (2001) Radical mechanisms of S-adenosylmethionine-dependent enzymes. Adv Protein Chem 58: 1–45 [DOI] [PubMed] [Google Scholar]
- Guldener U, Munsterkotter M, Kastenmuller G, Strack N, van Helden J, Lemer C, Richelles J, Wodak SJ, Garcia-Martinez J, Perez-Ortin JE, Michael H, Kaps A, Talla E, Dujon B, Andre B, Souciet JL, De Montigny J, Bon E, Gaillardin C, Mewes HW (2005) CYGD: the comprehensive yeast genome database. Nucleic Acids Res 33: D364–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield D, Feng YX, Lee BJ, Rein A, Levin JG, Oroszlan S (1989) Chromatographic analysis of the aminoacyl-tRNAs which are required for translation of codons at and around the ribosomal frameshift sites of HIV, HTLV-1, and BLV. Virology 173: 736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T (2006) Mechanistic insights into sulfur-relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol Cell 21: 97–108 [DOI] [PubMed] [Google Scholar]
- Kalhor HR, Luk K, Ramos A, Zobel-Thropp P, Clarke S (2001) Protein phosphatase methyltransferase 1 (Ppm1p) is the sole activity responsible for modification of the major forms of protein phosphatase 2A in yeast. Arch Biochem Biophys 395: 239–245 [DOI] [PubMed] [Google Scholar]
- Kalhor HR, Penjwini M, Clarke S (2005) A novel methyltransferase required for the formation of the hypermodified nucleoside wybutosine in eucaryotic tRNA. Biochem Biophys Res Commun 334: 433–440 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Suzuki T, Kapushoc ST, Rubio MA, Ghazvini J, Watanabe K, Simpson L, Suzuki T (2003) Wobble modification differences and subcellular localization of tRNAs in Leishmania tarentolae: implication for tRNA sorting mechanism. EMBO J 22: 657–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasai H, Goto M, Ikeda K, Zama M, Mizuno Y, Takemura S, Matsuura S, Sugimoto T, Goto T (1976) Structure of wye (Yt base) and wyosine (Yt) from Torulopsis utilis phenylalanine transfer ribonucleic acid. Biochemistry 15: 898–904 [DOI] [PubMed] [Google Scholar]
- Keith G, Dirheimer G (1980) Primary structure of Bombyx mori posterior silkgland tRNAPhe. Biochem Biophys Res Commun 92: 109–115 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Goto Y, Campos Y, Arenas J, Suzuki T (2005) Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc Natl Acad Sci USA 102: 7127–7132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Suzuki T (2004) Human mitochondrial diseases associated with tRNA wobble modification deficiency. RNA Biol 1: 145–148 [DOI] [PubMed] [Google Scholar]
- Konevega AL, Soboleva NG, Makhno VI, Semenkov YP, Wintermeyer W, Rodnina MV, Katunin VI (2004) Purine bases at position 37 of tRNA stabilize codon–anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. Rna 10: 90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y, Borek E, Grunberger D, Mushinski JF, Nishimura S (1982) Changes of post-transcriptional modification of wye base in tumor-specific tRNAPhe. Nucleic Acids Res 10: 6421–6432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, Cheung KH, Miller P, Gerstein M, Roeder GS, Snyder M (2002) Subcellular localization of the yeast proteome. Genes Dev 16: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TT, Agarwalla S, Stroud RM (2005) A unique RNA fold in the RumA–RNA–cofactor ternary complex contributes to substrate selectivity and enzymatic function. Cell 120: 599–611 [DOI] [PubMed] [Google Scholar]
- Leulliot N, Quevillon-Cheruel S, Sorel I, de La Sierra-Gallay IL, Collinet B, Graille M, Blondeau K, Bettache N, Poupon A, Janin J, van Tilbeurgh H (2004) Structure of protein phosphatase methyltransferase 1 (PPM1), a leucine carboxyl methyltransferase involved in the regulation of protein phosphatase 2A activity. J Biol Chem 279: 8351–8358 [DOI] [PubMed] [Google Scholar]
- Li HJ, Nakanishi K, Grunberger D, Weinstein IB (1973) Biosynthetic studies of the Y base in yeast phenylalanine tRNA. Incorporation of guanine. Biochem Biophys Res Commun 55: 818–823 [DOI] [PubMed] [Google Scholar]
- Munch HJ, Thiebe R (1975) Biosynthesis of the nucleoside Y in yeast tRNAPhe: incorporation of the 3-amino-3-carboxypropyl-group from methionine. FEBS Lett 51: 257–258 [DOI] [PubMed] [Google Scholar]
- Nakai Y, Umeda N, Suzuki T, Nakai M, Hayashi H, Watanabe K, Kagamiyama H (2004) Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J Biol Chem 279: 12363–12368 [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB Jr, Deerfield II DW (1997) GeneDoc: a tool for editing and annotating multiple sequence alignments. EMBNEW NEWS 4: 14 [Google Scholar]
- Niewmierzycka A, Clarke S (1999) S-adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J Biol Chem 274: 814–824 [DOI] [PubMed] [Google Scholar]
- Pergolizzi RG, Engelhardt DL, Grunberger D (1979) Incorporation of lysine into Y base of phenylalanine tRNA in Vero cells. Nucleic Acids Res 6: 2209–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierrel F, Douki T, Fontecave M, Atta M (2004) MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J Biol Chem 279: 47555–47563 [DOI] [PubMed] [Google Scholar]
- Rozenski J, Crain PF, McCloskey JA (1999) The RNA modification database: 1999 update. Nucleic Acids Res 27: 196–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res 29: 1097–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, Ote T, Kato J, Watanabe K, Sekine Y, Suzuki T (2003) An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol Cell 12: 689–698 [DOI] [PubMed] [Google Scholar]
- Suzuki T (2005) Biosynthesis and function of tRNA wobble modifications. In Fine-Tuning of RNA Functions by Modification and Editing, Vol. 12. pp 24–69. New York: Springer-Verlag [Google Scholar]
- Takano A, Endo T, Yoshihisa T (2005) tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309: 140–142 [DOI] [PubMed] [Google Scholar]
- Thiebe R, Poralla K (1973) Origin of the nucleoside Y in yeast tRNAPhe. FEBS Lett 38: 27–28 [DOI] [PubMed] [Google Scholar]
- Thiebe R, Zachau HG (1968) A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur J Biochem 5: 546–555 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakabayashi S, Kimura T, Fukuyama K, Matsubara H, Rogers LJ (1989) The amino acid sequence of a flavodoxin from the eukaryotic red alga Chondrus crispus. Biochem J 263: 981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Ashby DG, Moreno CS, Ogris E, Yeong FM, Corbett AH, Pallas DC (2001) Carboxymethylation of the PP2A catalytic subunit in Saccharomyces cerevisiae is required for efficient interaction with the B-type subunits Cdc55p and Rts1p. J Biol Chem 276: 1570–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Tolstykh T, Lee J, Boyd K, Stock JB, Broach JR (2000) Carboxyl methylation of the phosphoprotein phosphatase 2A catalytic subunit promotes its functional association with regulatory subunits in vivo. EMBO J 19: 5672–5681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T (2003) Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 14: 3266–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Sitaramaiah D, Noon KR, Guymon R, Hashizume T, McCloskey JA (2004a) Structures of two new ‘minimalist' modified nucleosides from archaeal tRNA. Bioorg Chem 32: 82–91 [DOI] [PubMed] [Google Scholar]
- Zhou S, Sitaramaiah D, Pomerantz SC, Crain PF, McCloskey JA (2004b) Tandem mass spectrometry for structure assignments of wye nucleosides from transfer RNA. Nucleos Nucl Nucleic Acids 23: 41–50 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


