Figure 3a.
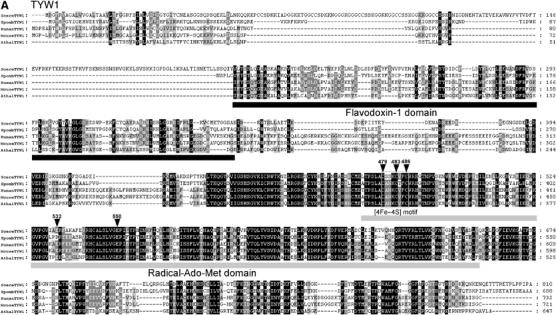
Sequence alignments of the TYW proteins. Each TYW protein is aligned with a set of protein homologs from S. cerevisiae (Scere), S. pombe (Spomb), Homo sapiens (Human), Mus musculus (Mouse), A. thaliana (Athal) and O. sativa (Osati). Multiple alignment of each sequence was carried out by Clustal X (Thompson et al, 1997) and displayed by Genedoc multiple sequence alignment editor (Nicholas et al, 1997). White letters in black boxes represent amino-acid residues identical in all species, while white letters in gray boxes represent residues with ∼80% homology. Black letters in gray boxes represent residues with ∼60% homology. (A) Sequence alignment of TYW1 with its homologs. Two conserved domains, flavodoxin-1 domain and Radical-Ado-Met domain, are underlined. The boxed region represents the [4Fe–4S] motif. Positions for site-directed mutagenesis are indicated by arrowheads.
