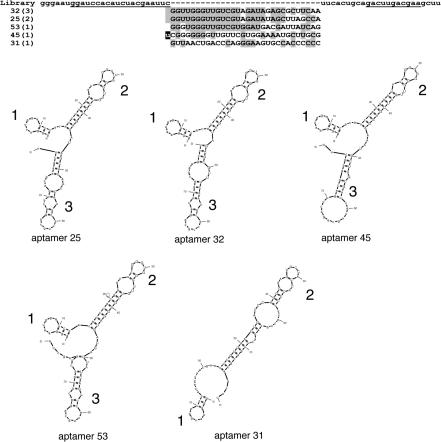
Figure 3.
Sequences and structure of selected aptamers. (Upper) Sequences of the primers, the RNA pool and the polβ aptamers. The PCR primer annealing sites within the constant regions are underlined. For the RNA library, the randomized region (n30) is flanked by the 5′ and 3′ constant regions. The names of the representative aptamers are shown on the left. The number in parentheses indicates the number of independent clones of the identical sequence. In aptamer 45, the last C in the 5′ constant region has been mutated to a U. The sequences are shown with the most similar sequences grouped together according to Clustal (49). (Lower) The Mfold program (available at http://www.bioinfo.rpi.edu/applications/mfold) was used to derive secondary structural models for aptamers 25 (−17.6 kcal/mol, next most stable −17.0 kcal/mol), 31 (−15.3 kcal/mol, next most stable −14.5 kcal/mol), 32 (−19.3 kcal/mol, next most stable −18.3 kcal/mol), 45 (−21.2 kcal/mol, next most stable −20.6 kcal/mol) and 53 (−20.2 kcal/mol, next most stable −19.4 kcal/mol). Only the most stable predicted structure is shown.